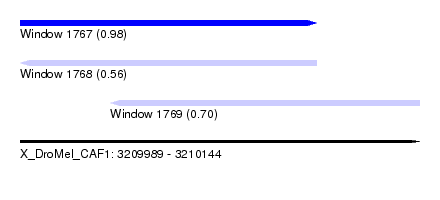
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,209,989 – 3,210,144 |
| Length | 155 |
| Max. P | 0.976168 |

| Location | 3,209,989 – 3,210,104 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 65.27 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -11.91 |
| Energy contribution | -13.60 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3209989 115 + 22224390 AAAUGUAUGCGAAGUGUUAGUCUGGUU-UUAUUUU---UUUUUGUUUUUUGCAUACCUUUUUGUUUAUUGUGUUUGAGAGCAAACACAAGCGUUAGCAAAUAAAAACGUAAAAAUAGCG ....((((((((((.............-.......---........)))))))))).(((((((((.(((((((((....)))))))))((....)))))))))))............. ( -23.99) >DroSec_CAF1 19373 92 + 1 --------GCGAAGUGUU--------------UUU---UUGUUUUUUUUUUCAUACCUUUUUGUUUAUUGUGUUUGAGAGCAAACACAAGCGUCAGCAAAUA--AACGUAAAAAUAGCG --------((....((((--------------(((---.(((((.(((...................(((((((((....)))))))))((....))))).)--)))).))))))))). ( -17.90) >DroEre_CAF1 21227 107 + 1 AAAUGUAUGCAAAGUGUUUGUUUUUUUU----------UUUUCUUUUUUUGCAUGCCUAUUUGUUUAUUGUGUUUGAGAGCAAACACAAGCGUCAGCAAAUA--AACGUAAAAAUAGCG ....((((((((((..............----------........))))))))))((((((((((((((((((((....)))))))..((....)).))))--)))....)))))).. ( -24.95) >DroAna_CAF1 32330 99 + 1 AAAUGUAUGCAGAGUAUUUUUUUUUUGCAUACUUCUUAUAGUG--UUUUUUCUCAUUUGUUUGUUUAGUUUGUUUGAGAGCAAACAUUA----CGACAAAAA--AAA------------ ....((((((((((........))))))))))...(..(((((--(((.((((((...................)))))).))))))))----..)......--...------------ ( -23.01) >consensus AAAUGUAUGCAAAGUGUUUGUUUUUUU_____UUU___UUGUUUUUUUUUGCAUACCUUUUUGUUUAUUGUGUUUGAGAGCAAACACAAGCGUCAGCAAAUA__AACGUAAAAAUAGCG ....((((((((((................................))))))))))...((((((..(((((((((....))))))))).....))))))................... (-11.91 = -13.60 + 1.69)

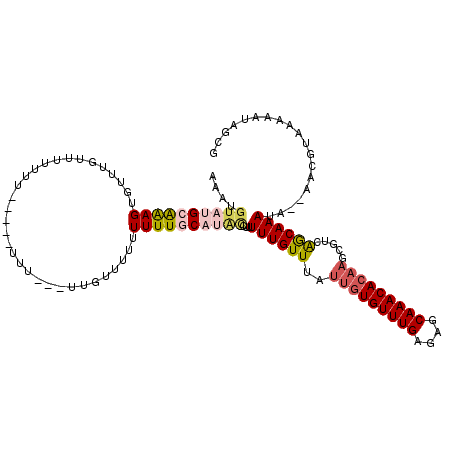
| Location | 3,209,989 – 3,210,104 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 65.27 |
| Mean single sequence MFE | -16.69 |
| Consensus MFE | -6.28 |
| Energy contribution | -7.03 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.561102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3209989 115 - 22224390 CGCUAUUUUUACGUUUUUAUUUGCUAACGCUUGUGUUUGCUCUCAAACACAAUAAACAAAAAGGUAUGCAAAAAACAAAAA---AAAAUAA-AACCAGACUAACACUUCGCAUACAUUU ............((((......((....))(((((((((....))))))))).))))......((((((............---.......-.................)))))).... ( -17.52) >DroSec_CAF1 19373 92 - 1 CGCUAUUUUUACGUU--UAUUUGCUGACGCUUGUGUUUGCUCUCAAACACAAUAAACAAAAAGGUAUGAAAAAAAAAACAA---AAA--------------AACACUUCGC-------- ...(((((((..(((--((((.((....))..(((((((....))))))))))))))..)))))))...............---...--------------..........-------- ( -13.40) >DroEre_CAF1 21227 107 - 1 CGCUAUUUUUACGUU--UAUUUGCUGACGCUUGUGUUUGCUCUCAAACACAAUAAACAAAUAGGCAUGCAAAAAAAGAAAA----------AAAAAAAACAAACACUUUGCAUACAUUU ............(((--((((((.......(((((((((....)))))))))....)))))))))(((((((.........----------...............)))))))...... ( -19.46) >DroAna_CAF1 32330 99 - 1 ------------UUU--UUUUUGUCG----UAAUGUUUGCUCUCAAACAAACUAAACAAACAAAUGAGAAAAAA--CACUAUAAGAAGUAUGCAAAAAAAAAAUACUCUGCAUACAUUU ------------..(--(((((.(((----(..((((((.................)))))).)))).))))))--...........(((((((..............))))))).... ( -16.37) >consensus CGCUAUUUUUACGUU__UAUUUGCUGACGCUUGUGUUUGCUCUCAAACACAAUAAACAAAAAGGUAUGAAAAAAAAAAAAA___AAA_____AAAAAAACAAACACUUCGCAUACAUUU ..............................(((((((((....)))))))))................................................................... ( -6.28 = -7.03 + 0.75)

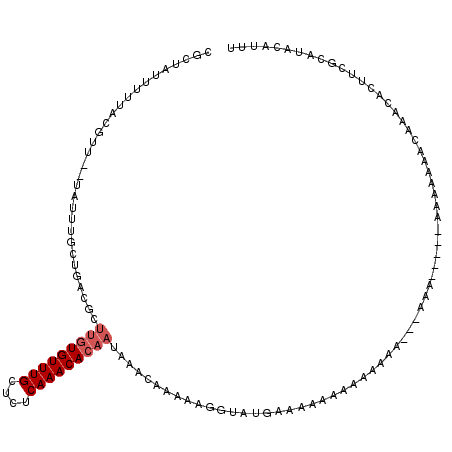
| Location | 3,210,024 – 3,210,144 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -18.09 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3210024 120 - 22224390 ACUAAUUAUGAAGCAAUUCCAUUGCAAUGCCAGCGAUCUUCGCUAUUUUUACGUUUUUAUUUGCUAACGCUUGUGUUUGCUCUCAAACACAAUAAACAAAAAGGUAUGCAAAAAACAAAA .........(((....)))..(((((.((((((((.....))))........((((......((....))(((((((((....))))))))).)))).....)))))))))......... ( -28.60) >DroSec_CAF1 19387 118 - 1 ACUAAUUAUGAAGCAAUUCCAUUGCAAUCCCCGCGAUCUUCGCUAUUUUUACGUU--UAUUUGCUGACGCUUGUGUUUGCUCUCAAACACAAUAAACAAAAAGGUAUGAAAAAAAAAACA .....(((((..(((((...))))).....(((((.....))).........(((--((((.((....))..(((((((....)))))))))))))).....)))))))........... ( -21.40) >DroEre_CAF1 21256 118 - 1 ACUAAUUAUCAAGCAAUUUCAUUGCAGUCCCAGUGAUCUUCGCUAUUUUUACGUU--UAUUUGCUGACGCUUGUGUUUGCUCUCAAACACAAUAAACAAAUAGGCAUGCAAAAAAAGAAA .....................(((((.....((((.....))))........(((--((((((.......(((((((((....)))))))))....))))))))).)))))......... ( -23.50) >DroYak_CAF1 24135 118 - 1 ACUAAUUAUGAAGCAAUUCCAUUGCAAUCCCAGCGGUCUUCGCUAUUUUUACGUU--UAUUUGCCGACGCUUGUGUUUGCUCUCAAACACAAUAAACAAAAAGGCAUGCAAAAAAAAAAA .........(((....)))..(((((...((((((.....))))........(((--((((.((....))..(((((((....)))))))))))))).....))..)))))......... ( -23.30) >consensus ACUAAUUAUGAAGCAAUUCCAUUGCAAUCCCAGCGAUCUUCGCUAUUUUUACGUU__UAUUUGCUGACGCUUGUGUUUGCUCUCAAACACAAUAAACAAAAAGGCAUGCAAAAAAAAAAA .....................(((((...((((((.....))))........(((.......((....))(((((((((....)))))))))..))).....))..)))))......... (-18.09 = -18.40 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:20 2006