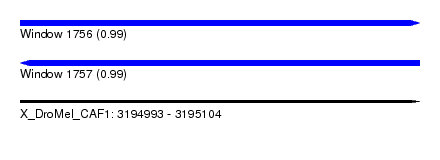
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,194,993 – 3,195,104 |
| Length | 111 |
| Max. P | 0.988758 |

| Location | 3,194,993 – 3,195,104 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -16.53 |
| Energy contribution | -17.75 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
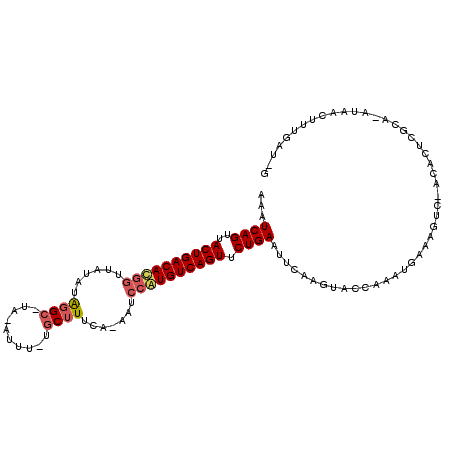
>X_DroMel_CAF1 3194993 111 + 22224390 AAAUCAGUUACUGACACGAUUAUAUACGCCUAAAUUUACGCUUUCAUAAUCGAUGUCAGUUCUGAAUUCAAGUACCAAAUGAAAGUCAACACUCGCAAAUAAUUUUGAUUG ...((((..((((((((((((((..(.((.((....)).)).)..))))))).))))))).))))..(((((.......((..(((....)))..))......)))))... ( -24.12) >DroSec_CAF1 4592 104 + 1 AAAUCAGUUACUGACAUGGUUAUAUAGGC----AUUUGUGCUUUCAAAAUCCAUGUCAGUUCUGAAUUCAAGUACCAAAUGAGAGUC-ACACUCGCA-AUAACUUUGAU-G ...((((..((((((((((......((((----(....))))).......)))))))))).))))..(((((.......((.(((..-...))).))-.....))))).-. ( -29.62) >DroYak_CAF1 6518 104 + 1 AACUCAGUUACUGACACGGUAG--GGGGCAUAGAUUU-UCCUUUCA-AAUCCGUGUCAGUUCUGAGUUCAAGUACCAAAUGAAACUC-AAACUCGUA-AUAACUGUGUU-G (((.(((((((((((((((.((--((((........)-)))))...-...)))))))))....(((((..(((..(....)..))).-.)))))...-.)))))).)))-. ( -32.50) >consensus AAAUCAGUUACUGACACGGUUAUAUAGGC_UA_AUUU_UGCUUUCA_AAUCCAUGUCAGUUCUGAAUUCAAGUACCAAAUGAAAGUC_ACACUCGCA_AUAACUUUGAU_G ...((((..((((((((((......((((..........)))).......)))))))))).)))).............................................. (-16.53 = -17.75 + 1.23)

| Location | 3,194,993 – 3,195,104 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
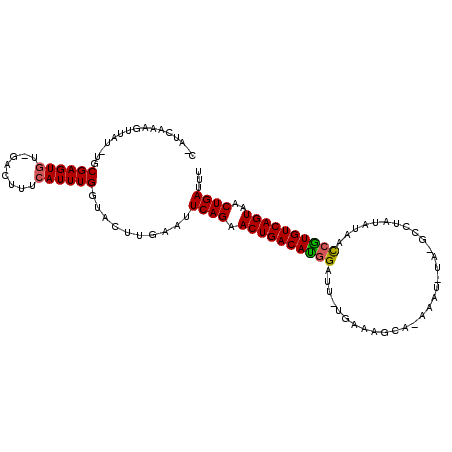
>X_DroMel_CAF1 3194993 111 - 22224390 CAAUCAAAAUUAUUUGCGAGUGUUGACUUUCAUUUGGUACUUGAAUUCAGAACUGACAUCGAUUAUGAAAGCGUAAAUUUAGGCGUAUAUAAUCGUGUCAGUAACUGAUUU ...((((.((((..((.((((....)))).))..))))..))))..((((.(((((((.((((((((...((.((....)).))...)))))))))))))))..))))... ( -30.50) >DroSec_CAF1 4592 104 - 1 C-AUCAAAGUUAU-UGCGAGUGU-GACUCUCAUUUGGUACUUGAAUUCAGAACUGACAUGGAUUUUGAAAGCACAAAU----GCCUAUAUAACCAUGUCAGUAACUGAUUU .-.((((.((...-((.(((...-..))).))......))))))..((((.((((((((((.((......(((....)----))......))))))))))))..))))... ( -25.40) >DroYak_CAF1 6518 104 - 1 C-AACACAGUUAU-UACGAGUUU-GAGUUUCAUUUGGUACUUGAACUCAGAACUGACACGGAUU-UGAAAGGA-AAAUCUAUGCCCC--CUACCGUGUCAGUAACUGAGUU .-(((.(((((..-...(((((.-.(((.((....)).)))..)))))...((((((((((...-....(((.-............)--)).))))))))))))))).))) ( -34.02) >consensus C_AUCAAAGUUAU_UGCGAGUGU_GACUUUCAUUUGGUACUUGAAUUCAGAACUGACAUGGAUU_UGAAAGCA_AAAU_UA_GCCUAUAUAACCGUGUCAGUAACUGAUUU ................((((((........))))))..........((((.((((((((((...............................))))))))))..))))... (-18.14 = -18.15 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:09 2006