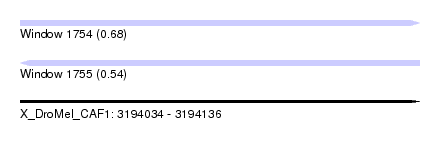
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,194,034 – 3,194,136 |
| Length | 102 |
| Max. P | 0.679169 |

| Location | 3,194,034 – 3,194,136 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.12 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3194034 102 + 22224390 GCGGGGCGCUGUUCGCCGCUUGCAUACACGAUGUUGAUCAUCCCGGCUUAACCAAUCAGUUCUUGGUUAACUCAAGUGCGU----AUUACCU--ACA--CUUUAGUUCGU ..((((((.....))))....((((....((((.....))))..(..((((((((.......))))))))..)..))))..----....)).--...--........... ( -22.70) >DroVir_CAF1 5676 99 + 1 GCGGGGCGCUGUUCGCUGCUUGUAUACACGAUGUUGAUCAUCCUGGUUUAACCAAUCAAUUCUUGGUUAAUUCAAGUGAGU----AACAGAA--AUA--C--CU-UUAAA ..(((...(((((.((((((((....((.((((.....)))).))..((((((((.......))))))))..))))).)))----)))))..--...--)--))-..... ( -24.80) >DroPse_CAF1 4991 96 + 1 GCGGGGCACUGUUUGCCGCUUGCAUACACGAUGUUGAUCAUCCAGGAUUAACCAAUCAGUUUUUGGUUAACUCAAGUGAGU----AUCUCUC----------CUGGUAUU ((((((((.....)))).).)))((((..((((.....))))(((((.(((((((.......)))))))((((....))))----.....))----------))))))). ( -28.80) >DroGri_CAF1 4395 101 + 1 GCGGGGCCCUGUUCGCCGCUUGUAUUCACGAUGUUGAUCAUCCUGGCUUAACCAAUCAGUUUUUGGUUAAUUCAAGUACGAACGAAACACAA--ACA------U-UUCAA ((((.(.......).))))(((((((...((((.....)))).((..((((((((.......))))))))..)))))))))..((((.....--...------)-))).. ( -23.00) >DroWil_CAF1 4128 106 + 1 GCGGUGCAUUGUUCGCCGCUUGUAUACACGAUGUUGAUCAUCCUGGCUUAACCAAUCAGUUUUUGGUUAACUCAAGUGAGU----CUUUUGCGUACAGACAGCAAAUCGA ((((((.......))))))((((((.((.((((.....)))).))((.(((((((.......)))))))((((....))))----.....))))))))............ ( -27.90) >DroPer_CAF1 5125 96 + 1 GCGGGGCACUGUUUGCCGCUUGCAUACACGAUGUUGAUCAUCCAGGAUUAACCAAUCAGUUUUUGGUUAACUCAAGUGAGU----AUCUCUC----------CUGGUAUU ((((((((.....)))).).)))((((..((((.....))))(((((.(((((((.......)))))))((((....))))----.....))----------))))))). ( -28.80) >consensus GCGGGGCACUGUUCGCCGCUUGCAUACACGAUGUUGAUCAUCCUGGCUUAACCAAUCAGUUUUUGGUUAACUCAAGUGAGU____AUCACAC__ACA_____CUGUUAAA ((((.(.......).)))).......(((((((.....))))..(..((((((((.......))))))))..)..)))................................ (-19.31 = -19.12 + -0.19)

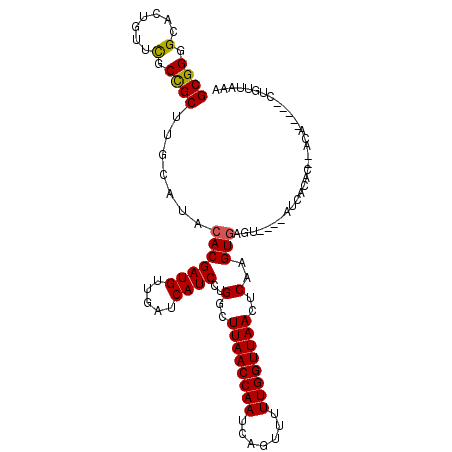
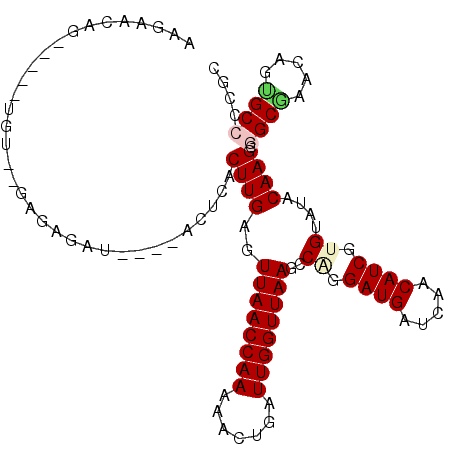
| Location | 3,194,034 – 3,194,136 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -15.63 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3194034 102 - 22224390 ACGAACUAAAG--UGU--AGGUAAU----ACGCACUUGAGUUAACCAAGAACUGAUUGGUUAAGCCGGGAUGAUCAACAUCGUGUAUGCAAGCGGCGAACAGCGCCCCGC .((........--.((--..(((.(----((((.((((..((((((((.......))))))))..))))..(((....)))))))))))..))((((.....)))).)). ( -25.80) >DroVir_CAF1 5676 99 - 1 UUUAA-AG--G--UAU--UUCUGUU----ACUCACUUGAAUUAACCAAGAAUUGAUUGGUUAAACCAGGAUGAUCAACAUCGUGUAUACAAGCAGCGAACAGCGCCCCGC .....-.(--(--(..--..(((((----.....((((..((((((((.......))))))))..((.((((.....)))).))....)))).....))))).))).... ( -22.60) >DroPse_CAF1 4991 96 - 1 AAUACCAG----------GAGAGAU----ACUCACUUGAGUUAACCAAAAACUGAUUGGUUAAUCCUGGAUGAUCAACAUCGUGUAUGCAAGCGGCAAACAGUGCCCCGC ....((((----------((.....----((((....))))(((((((.......))))))).))))))(((((....)))))........(.((((.....)))).).. ( -25.40) >DroGri_CAF1 4395 101 - 1 UUGAA-A------UGU--UUGUGUUUCGUUCGUACUUGAAUUAACCAAAAACUGAUUGGUUAAGCCAGGAUGAUCAACAUCGUGAAUACAAGCGGCGAACAGGGCCCCGC .....-.------.((--((((((((((.((((.((((..((((((((.......))))))))..)))))))).......)).))))))))))(((.......))).... ( -27.60) >DroWil_CAF1 4128 106 - 1 UCGAUUUGCUGUCUGUACGCAAAAG----ACUCACUUGAGUUAACCAAAAACUGAUUGGUUAAGCCAGGAUGAUCAACAUCGUGUAUACAAGCGGCGAACAAUGCACCGC ..(.(((((((..((((..((...(----(.(((((((..((((((((.......))))))))..)))).))))).......))..))))..)))))))).......... ( -29.30) >DroPer_CAF1 5125 96 - 1 AAUACCAG----------GAGAGAU----ACUCACUUGAGUUAACCAAAAACUGAUUGGUUAAUCCUGGAUGAUCAACAUCGUGUAUGCAAGCGGCAAACAGUGCCCCGC ....((((----------((.....----((((....))))(((((((.......))))))).))))))(((((....)))))........(.((((.....)))).).. ( -25.40) >consensus AAGAACAG_____UGU__GAGAGAU____ACUCACUUGAGUUAACCAAAAACUGAUUGGUUAAGCCAGGAUGAUCAACAUCGUGUAUACAAGCGGCGAACAGUGCCCCGC ..................................((((..((((((((.......))))))))..((.((((.....)))).))....)))).((((.....)))).... (-15.63 = -16.05 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:07 2006