| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,149,841 – 3,149,932 |
| Length | 91 |
| Max. P | 0.878724 |

| Location | 3,149,841 – 3,149,932 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 93.36 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -19.80 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
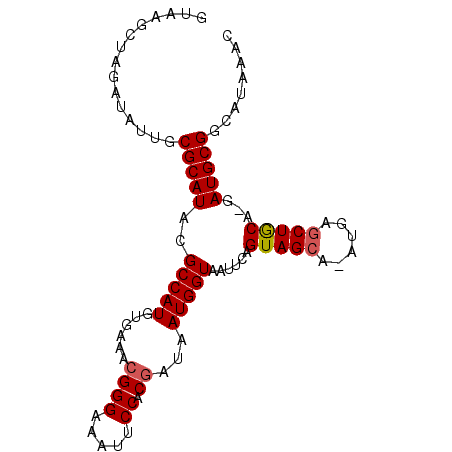
>X_DroMel_CAF1 3149841 91 + 22224390 GUAAGCUAGAUAUUGCGCAUACGCCAUGUGAAACGGGAAAUUCCACAAUAAUGGUAAUUCAGUAGCA-AUGAGCUACA-GAUGCGGCAUAAAC .............(((((((..(((((........((.....))......)))))......(((((.-....))))).-.)))).)))..... ( -20.74) >DroSec_CAF1 107380 91 + 1 GUAAGCUAGAUAUUUCGCAUACGCCAUGUGAAACGGGAAAUUCCACGAUAAUGGUGAUUCAGUAGCA-AUGAGCUGCA-GAUGCGGCAUAAAU ..........(((.((((((.((((((......((((.....)).))...)))))).....(((((.-....))))).-.)))))).)))... ( -24.80) >DroSim_CAF1 108577 91 + 1 GUAAGCUAGAUAUUGCGCAUACGCCAUGUGAAGCGGGAAAUUCCACGAUAAUGGUGAUUCAGUAGCA-AUGAGCUGCA-GAUGCGGCAUAAAC .............(((((((.((((((......((((.....)).))...)))))).....(((((.-....))))).-.)))).)))..... ( -25.10) >DroEre_CAF1 113940 92 + 1 GGAAGCUACAUAUUGCGCAUACGCCAUGUGAAACGGGAAAUUCCACGGUAAUGGUAAUUCAGUAGCAAAUGAGCUGCA-GAUGCGGCAUAAAC ....(((((..(((((.(((..(((.((.(((........))))).))).))))))))...)))))......(((((.-...)))))...... ( -25.80) >DroYak_CAF1 115269 92 + 1 GGAAGCUAAAUAUUGCGCAUACGCCAUGUGAAACGGGAAAUUCCACGAUAAUGGUAAUUCAGUAGCA-AUGAACUACAAAAUGCGGCAUAAAC .............(((((((..(((((......((((.....)).))...)))))......((((..-.....))))...)))).)))..... ( -17.30) >consensus GUAAGCUAGAUAUUGCGCAUACGCCAUGUGAAACGGGAAAUUCCACGAUAAUGGUAAUUCAGUAGCA_AUGAGCUGCA_GAUGCGGCAUAAAC ...............(((((..(((((......((((.....)).))...)))))......(((((......)))))...)))))........ (-19.80 = -19.96 + 0.16)

| Location | 3,149,841 – 3,149,932 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 93.36 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -17.22 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
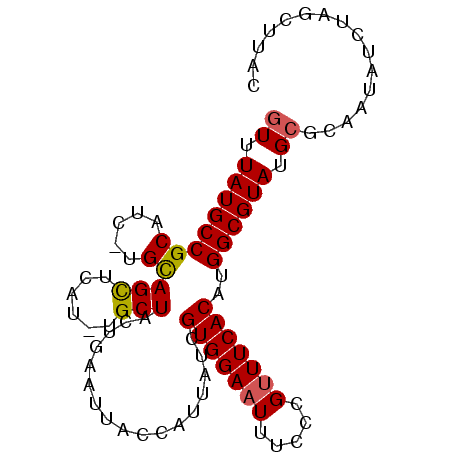
>X_DroMel_CAF1 3149841 91 - 22224390 GUUUAUGCCGCAUC-UGUAGCUCAU-UGCUACUGAAUUACCAUUAUUGUGGAAUUUCCCGUUUCACAUGGCGUAUGCGCAAUAUCUAGCUUAC ((.(((((((..((-.(((((....-.))))).))...........((((((((.....))))))))))))))).))((........)).... ( -25.50) >DroSec_CAF1 107380 91 - 1 AUUUAUGCCGCAUC-UGCAGCUCAU-UGCUACUGAAUCACCAUUAUCGUGGAAUUUCCCGUUUCACAUGGCGUAUGCGAAAUAUCUAGCUUAC ...(((..(((((.-((((((....-.))).................(((((((.....)))))))...))).)))))..))).......... ( -19.40) >DroSim_CAF1 108577 91 - 1 GUUUAUGCCGCAUC-UGCAGCUCAU-UGCUACUGAAUCACCAUUAUCGUGGAAUUUCCCGCUUCACAUGGCGUAUGCGCAAUAUCUAGCUUAC ((.((((((((...-.))(((....-.))).................((((......)))).......)))))).))((........)).... ( -19.60) >DroEre_CAF1 113940 92 - 1 GUUUAUGCCGCAUC-UGCAGCUCAUUUGCUACUGAAUUACCAUUACCGUGGAAUUUCCCGUUUCACAUGGCGUAUGCGCAAUAUGUAGCUUCC ((.((((((((...-.))(((......))).................(((((((.....)))))))..)))))).))((........)).... ( -20.90) >DroYak_CAF1 115269 92 - 1 GUUUAUGCCGCAUUUUGUAGUUCAU-UGCUACUGAAUUACCAUUAUCGUGGAAUUUCCCGUUUCACAUGGCGUAUGCGCAAUAUUUAGCUUCC ((.(((((((......((((((((.-......)))))))).......(((((((.....))))))).))))))).))((........)).... ( -23.20) >consensus GUUUAUGCCGCAUC_UGCAGCUCAU_UGCUACUGAAUUACCAUUAUCGUGGAAUUUCCCGUUUCACAUGGCGUAUGCGCAAUAUCUAGCUUAC ((.((((((((.....))(((......))).................(((((((.....)))))))..)))))).))................ (-17.22 = -17.22 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:56 2006