
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,127,197 – 3,127,349 |
| Length | 152 |
| Max. P | 0.999143 |

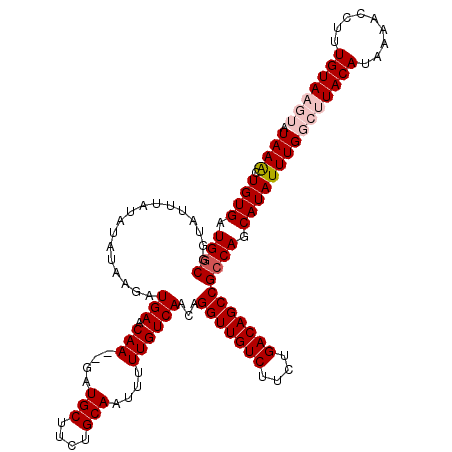
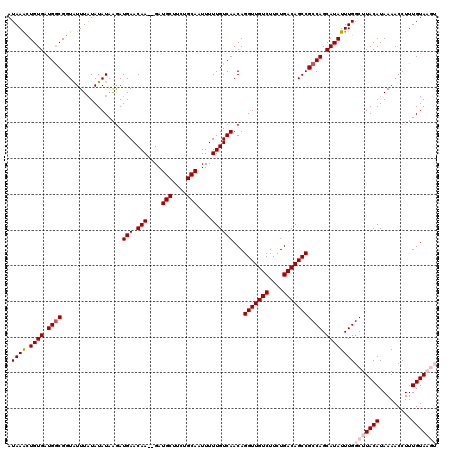
| Location | 3,127,197 – 3,127,317 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -28.16 |
| Energy contribution | -29.40 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3127197 120 + 22224390 AUAAACUGUGAUGACGGUAUUUUUAUAUAAGAUGAACAAGAGAUGCUUCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAAGU ....(((((....)))))...((((((.(((.(((.((((((.(((....))).)))))))))...(((((((....)))))))(((((.....)))))...........))))))))). ( -31.60) >DroSec_CAF1 85527 116 + 1 AUAAACUGUGAUGGCGGUAUUUAUAUAUAAGAUGAACAA--GAUGCUCCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAU-- .((((.((((.((((...............(.(((.(((--(((((....)))..)))))))).).(((((((....))))))))))).))))))))...((((.........)))).-- ( -29.00) >DroSim_CAF1 86771 116 + 1 AUAAACUGUGAUGGCGGUAUUUAUAUAUAAGAUGAACAA--GAUGCUCCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAU-- .((((.((((.((((...............(.(((.(((--(((((....)))..)))))))).).(((((((....))))))))))).))))))))...((((.........)))).-- ( -29.00) >DroEre_CAF1 90098 118 + 1 AUAAGUUGUGCUGGCGGUAUUUAUAUAUGGGAUGAACAA--GAUGCUUCUGCAGUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAAGU .(((((.((((((((.................(((.(((--(((((....)))..))))))))...(((((((....))))))))))))))))))))(((((((.........))))))) ( -40.50) >DroYak_CAF1 92360 118 + 1 AUAAAUUGUGAUGGCGGUGUUUAUAUAUAAAAUGAACAA--GAUGCUUCUGCAGUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAAGU .(((((.(((.((((.................(((.(((--(((((....)))..))))))))...(((((((....))))))))))).))))))))(((((((.........))))))) ( -33.90) >consensus AUAAACUGUGAUGGCGGUAUUUAUAUAUAAGAUGAACAA__GAUGCUUCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAAGU .((((.((((.((((.................(((.(((....(((....)))....))))))...(((((((....))))))))))).))))))))(((((((.........))))))) (-28.16 = -29.40 + 1.24)

| Location | 3,127,197 – 3,127,317 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -24.10 |
| Energy contribution | -23.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3127197 120 - 22224390 ACUUACAAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGAAGCAUCUCUUGUUCAUCUUAUAUAAAAAUACCGUCAUCACAGUUUAU .........((((((((((((((((.......))))((.((((....)))).))....(((((....(((....)))....)))))....)))))))))...)))............... ( -23.70) >DroSec_CAF1 85527 116 - 1 --AUACAAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGGAGCAUC--UUGUUCAUCUUAUAUAUAAAUACCGCCAUCACAGUUUAU --.......(((((.....))))).....((.((((((.((((((.((.....)).))))))......(((((.....)--))))..................)))))).))........ ( -26.20) >DroSim_CAF1 86771 116 - 1 --AUACAAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGGAGCAUC--UUGUUCAUCUUAUAUAUAAAUACCGCCAUCACAGUUUAU --.......(((((.....))))).....((.((((((.((((((.((.....)).))))))......(((((.....)--))))..................)))))).))........ ( -26.20) >DroEre_CAF1 90098 118 - 1 ACUUACAAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAACUGCAGAAGCAUC--UUGUUCAUCCCAUAUAUAAAUACCGCCAGCACAACUUAU .........(((((.....))))).....(((((((((.((((((.((.....)).)))))).....(((....)))..--......................)))))))))........ ( -30.80) >DroYak_CAF1 92360 118 - 1 ACUUACAAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAACUGCAGAAGCAUC--UUGUUCAUUUUAUAUAUAAACACCGCCAUCACAAUUUAU .........(((((.....))))).....((.((((((.((((((.((.....)).)))))).....(((....)))..--......................)))))).))........ ( -24.20) >consensus ACUUACAAAGGUUUUAUGUAAGCCAAAUAUGCUGGCGGCUGUCAGAAGACAACCUGUUGACAAAAAUUGCAGAAGCAUC__UUGUUCAUCUUAUAUAUAAAUACCGCCAUCACAGUUUAU .........(((((.....))))).....((.((((((.((((....)))).......(((((....(((....)))....))))).................)))))).))........ (-24.10 = -23.94 + -0.16)

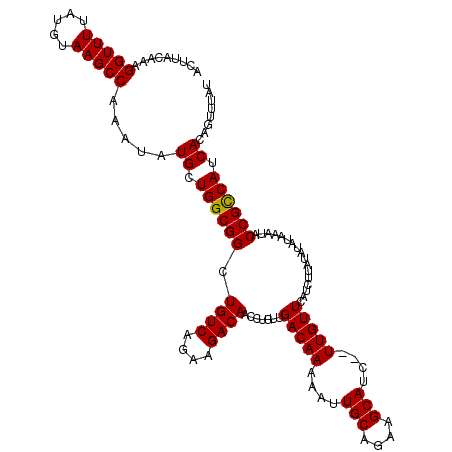
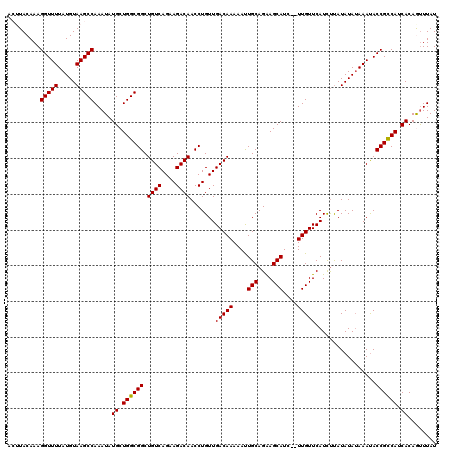
| Location | 3,127,237 – 3,127,349 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -26.80 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3127237 112 + 22224390 AGAUGCUUCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAAGUUGGCAUAAAGACGGUGGUA-AACAUGUCCACAG ...(((....))).(((.(((((((.(((((((....)))))))(((((.....)))))(((((.........)))))))))))).)))....((((..-.......)))).. ( -34.50) >DroSec_CAF1 85566 104 + 1 -GAUGCUCCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAU-------AAAAGACGGUGGUA-AACAUGUCCACAG -..(((....))).....((((....(((((((....)))))))(((((.....))))).((((.........)))).-------....))))((((..-.......)))).. ( -27.80) >DroSim_CAF1 86810 104 + 1 -GAUGCUCCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAU-------AAAAGACGGUGGUA-AACAUGUCCACAG -..(((....))).....((((....(((((((....)))))))(((((.....))))).((((.........)))).-------....))))((((..-.......)))).. ( -27.80) >DroEre_CAF1 90137 112 + 1 -GAUGCUUCUGCAGUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAAGUUGGCAUAAAGAGGGUGGUAAAACACGUCCACAG -....(((((........(((((((.(((((((....)))))))(((((.....)))))(((((.........))))))))))))...)))))((((..........)))).. ( -35.00) >DroYak_CAF1 92399 111 + 1 -GAUGCUUCUGCAGUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAAGUUGGCAUAAAGACGGUGGUA-AACACGUCCACUG -..(((....)))((((((((((((.(((((((....)))))))(((((.....)))))(((((.........))))))))))))..)))))(((((..-.......))))). ( -37.60) >consensus _GAUGCUUCUGCAAUUUUUGUCAACAGGUUGUCUUCUGACAGCCGCCAGCAUAUUUGGCUUACAUAAAACCUUUGUAAGUUGGCAUAAAGACGGUGGUA_AACAUGUCCACAG ...(((....))).....((((....(((((((....)))))))(((((.....))))).((((.........))))............))))((((..........)))).. (-26.80 = -27.00 + 0.20)

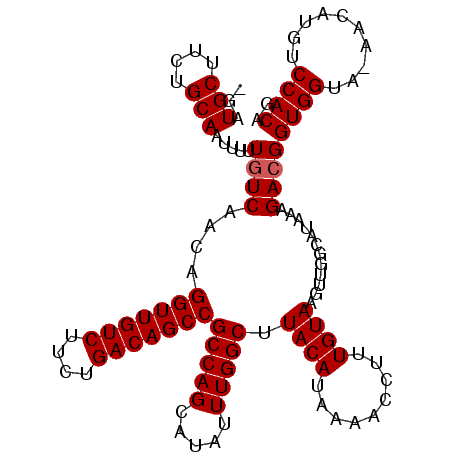
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:52 2006