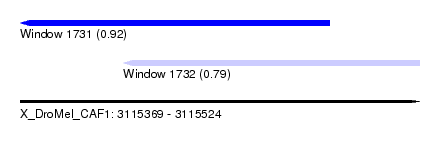
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,115,369 – 3,115,524 |
| Length | 155 |
| Max. P | 0.916231 |

| Location | 3,115,369 – 3,115,489 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.39 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.59 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
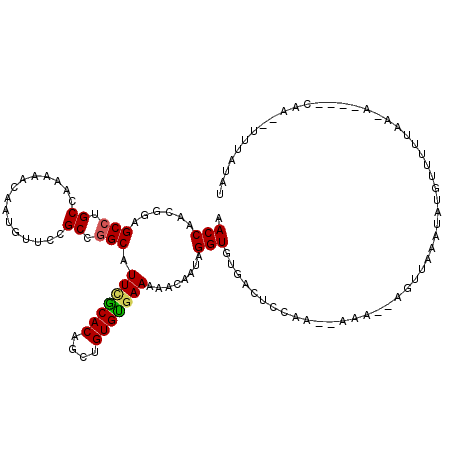
>X_DroMel_CAF1 3115369 120 - 22224390 AACCAAUGAAGCCUGCCAAAAACAAUAUUCCGCCGGCAUUCGCACAGUUGUGCGAAAAACACUAGGUGUGCUUUCAAACAAAUAAGUUAAUUAUGUUUUUUAUAGUGGCAAUUUUUAUGU .....((((((..(((((((((((.......(..(((.(((((((....)))))))..((((...)))))))..).(((......))).....))))))......)))))..)))))).. ( -25.50) >DroEre_CAF1 75100 118 - 1 AACCAACGGAGCCUGCCAAAAACAAUGUUCCGCCGGCAUUCGCACAGCUGUGUUAAAAACAAUAGGUGUAACUCCAAGUAAAGCAGUUAAAUAUGUUUUUAUCACAUCCAA--UUUAUAU .......((((((.((...............)).)))...........((((.((((((((.((....(((((.(.......).)))))..)))))))))).)))))))..--....... ( -21.46) >DroYak_CAF1 79129 97 - 1 AACCAACGGAGCGUGCCAAAAACAAUGUUCCGCCGGCAUUUACACAACUGUGUGAAACACAAUAGGUGUGGCGGC-------------AAAUCUGAUUUAAC--------A--UUUAUAU .......(((((((..........)))))))((((.(.(((((((....)))))))((((.....))))).))))-------------..............--------.--....... ( -28.10) >consensus AACCAACGGAGCCUGCCAAAAACAAUGUUCCGCCGGCAUUCGCACAGCUGUGUGAAAAACAAUAGGUGUGACUCCAA__AAA__AGUUAAAUAUGUUUUUAA_A____CAA__UUUAUAU .(((......(((.((...............)).))).(((((((....)))))))........)))..................................................... (-14.48 = -14.59 + 0.11)

| Location | 3,115,409 – 3,115,524 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -18.24 |
| Energy contribution | -19.32 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
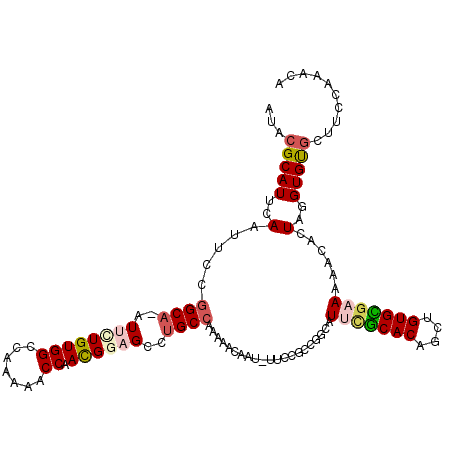
>X_DroMel_CAF1 3115409 115 - 22224390 AUACGCAUUCAAUUCGUGCAAAUUCUGUGGCCAAAAACCAAUGAAGCCUGCCAAAAACAAUAUUCCGCCGGCAUUCGCACAGUUGUGCGAAAAACACUAGGUGUGCUUUCAAACA ....(((..(((((.((((...(((..(((.......)))..)))(((.((...............)).)))....))))))))))))((((.((((...))))..))))..... ( -25.76) >DroSec_CAF1 74134 104 - 1 AUACGCAUUCAAUUCCGGCA-AUUCUGUGGCCAAAAACCAACGGAGCCUGCCAAAAACAU----------ACGUACGCACAGCUGUGCGCAAGACACUAGGUGUGCUUCCAAACG ................((((-.((((((((.......)).))))))..))))........----------......(((((.((((((....).))).)).)))))......... ( -29.30) >DroSim_CAF1 74785 104 - 1 AUACGCAUUCAAUUCCGGCA-AUUCUGUGGCCAAAAACCAACGGAGCCUGCCAAAAACAU----------ACGUACGCACAGCUGUGCGCAAGCUACUAGGUGCGCUUUCAAACG ....((..........((((-.((((((((.......)).))))))..))))........----------......((((....((((....)).))...))))))......... ( -27.70) >DroEre_CAF1 75138 114 - 1 AUACGCAUUCAAUUCCGGCC-AUAAUGUGGCCAAAAACCAACGGAGCCUGCCAAAAACAAUGUUCCGCCGGCAUUCGCACAGCUGUGUUAAAAACAAUAGGUGUAACUCCAAGUA .(((((..........((((-((...)))))).....((..((((((.((.......))..))))))..)).....))(((.((((((.....)).)))).)))........))) ( -26.40) >DroYak_CAF1 79151 109 - 1 AUACGCAUUCAAUUCCGGCA-AUAUUGUGGCCAAAAACCAACGGAGCGUGCCAAAAACAAUGUUCCGCCGGCAUUUACACAACUGUGUGAAACACAAUAGGUGUGGCGGC----- ...(((........(((((.-......(((.......)))..(((((((..........))))))))))))..(((((((....))))))).((((.....)))))))..----- ( -33.60) >consensus AUACGCAUUCAAUUCCGGCA_AUUCUGUGGCCAAAAACCAACGGAGCCUGCCAAAAACAAU_UUCCGCCGGCAUUCGCACAGCUGUGCGAAAAACACUAGGUGUGCUUCCAAACA ...(((((..(.....((((..((((((((.......)).))))))..)))).....................(((((((....)))))))......)..))))).......... (-18.24 = -19.32 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:46 2006