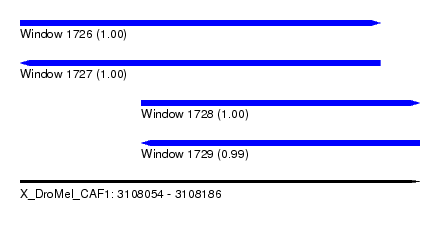
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,108,054 – 3,108,186 |
| Length | 132 |
| Max. P | 0.999996 |

| Location | 3,108,054 – 3,108,173 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.53 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -13.40 |
| Energy contribution | -15.40 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.52 |
| SVM decision value | 4.19 |
| SVM RNA-class probability | 0.999830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3108054 119 + 22224390 UGUUCGUUUUUUUUUUUUUUUUUGUAGGGUUGCGUUCGUGCGAAUUAUGUCAAUUGGGAUGCAAUGGUUCGAAAGUACAUAAUGACUUGGCAUUUUGCCAAGAGCCC-CCAAUUGCAUAA ....................((((((.((......)).))))))(((((.((((((((..((.(((...(....)..))).....((((((.....)))))).)).)-)))))))))))) ( -35.00) >DroSec_CAF1 67178 91 + 1 ----------UUUCGUUUUUUUUUUAAGGUUGCGUUCGUGCGAACUAUGUCAAUU------------------AGUACAUAAUGACUUGGCAUUUUGCCAAGAGCAA-CCAAUUGCAUAC ----------.................((((((((((....))))((((((....------------------.).)))))....((((((.....)))))).))))-)).......... ( -25.30) >DroYak_CAF1 72087 94 + 1 UU------------------UUUUCCACUUUAC------GCGUAUUAUGUCAAUUAGGAAGCAAUGAUUCGAAAAUACAUAAUUACUUGGCAUUUUGCCAAGACC--CCCAAUUGCAUAC ..------------------.............------(((((((...(((((((........))))).)).))))).......((((((.....))))))...--.......)).... ( -17.10) >consensus U_________UUU__UUUUUUUUUUAAGGUUGCGUUCGUGCGAAUUAUGUCAAUU_GGA_GCAAUG_UUCGAAAGUACAUAAUGACUUGGCAUUUUGCCAAGAGC___CCAAUUGCAUAC .....................................(((((((((((((......((((......))))......)))))))..((((((.....))))))..........)))))).. (-13.40 = -15.40 + 2.00)

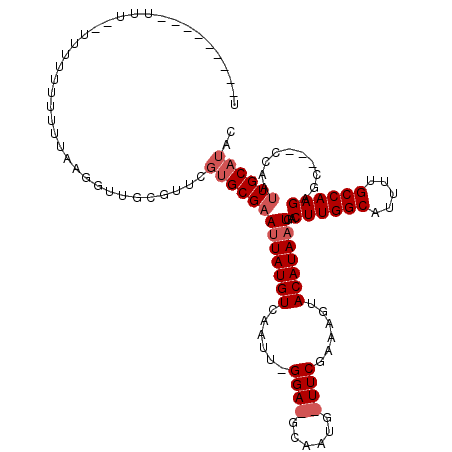
| Location | 3,108,054 – 3,108,173 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.53 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -13.74 |
| Energy contribution | -13.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.55 |
| SVM decision value | 6.08 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3108054 119 - 22224390 UUAUGCAAUUGG-GGGCUCUUGGCAAAAUGCCAAGUCAUUAUGUACUUUCGAACCAUUGCAUCCCAAUUGACAUAAUUCGCACGAACGCAACCCUACAAAAAAAAAAAAAAAAACGAACA ((((((((((((-((((.((((((.....))))))......((......)).......)).))))))))).)))))...((......))............................... ( -26.90) >DroSec_CAF1 67178 91 - 1 GUAUGCAAUUGG-UUGCUCUUGGCAAAAUGCCAAGUCAUUAUGUACU------------------AAUUGACAUAGUUCGCACGAACGCAACCUUAAAAAAAAAACGAAA---------- ..........((-((((.((((((.....))))))....(((((...------------------.....)))))((((....)))))))))).................---------- ( -24.30) >DroYak_CAF1 72087 94 - 1 GUAUGCAAUUGGG--GGUCUUGGCAAAAUGCCAAGUAAUUAUGUAUUUUCGAAUCAUUGCUUCCUAAUUGACAUAAUACGC------GUAAAGUGGAAAA------------------AA .((((((((((((--((.((((((.....)))))).......(((............))))))))))))).))))...(((------.....))).....------------------.. ( -23.70) >consensus GUAUGCAAUUGG___GCUCUUGGCAAAAUGCCAAGUCAUUAUGUACUUUCGAA_CAUUGC_UCC_AAUUGACAUAAUUCGCACGAACGCAACCUUAAAAAAAAAA__AAA_________A ...(((............((((((.....))))))..(((((((..........................)))))))..........))).............................. (-13.74 = -13.30 + -0.44)

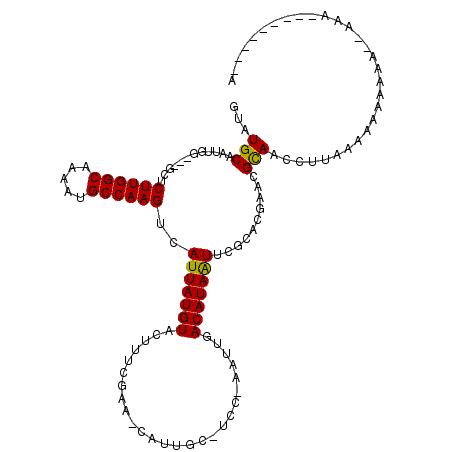
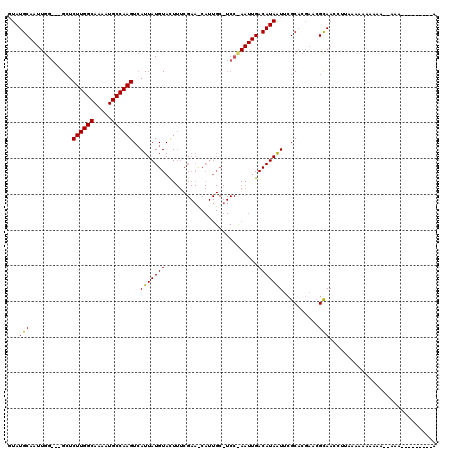
| Location | 3,108,094 – 3,108,186 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 74.46 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -11.95 |
| Energy contribution | -12.45 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3108094 92 + 22224390 CGAAUUAUGUCAAUUGGGAUGCAAUGGUUCGAAAGUACAUAAUGACUUGGCAUUUUGCCAAGAGCCC-CCAAUUGCAUAAAUGUGCAUAAAUU ....(((((.((((((((..((.(((...(....)..))).....((((((.....)))))).)).)-))))))))))))............. ( -30.20) >DroSec_CAF1 67208 74 + 1 CGAACUAUGUCAAUU------------------AGUACAUAAUGACUUGGCAUUUUGCCAAGAGCAA-CCAAUUGCAUACAUGUGCAUAAAUU ...............------------------.((((((.....((((((.....)))))).((((-....))))....))))))....... ( -19.10) >DroEre_CAF1 67753 78 + 1 CGAAUUAUGACAAUUAGGAAGCAAUGAUUCGA---------------UGGCAUUUCGCCAAGAGCCCCCAAAUUGCAUACAUGUGCAUAAAUU ((((((((...............)))))))).---------------((((.....)))).............(((((....)))))...... ( -15.26) >DroYak_CAF1 72103 91 + 1 CGUAUUAUGUCAAUUAGGAAGCAAUGAUUCGAAAAUACAUAAUUACUUGGCAUUUUGCCAAGACC--CCCAAUUGCAUACAUGUGCAUAAAUU .((((((((.(((((.((.....((((((....))).))).....((((((.....))))))...--)).)))))))))...))))....... ( -19.30) >consensus CGAAUUAUGUCAAUUAGGAAGCAAUGAUUCGA_AGUACAUAAUGACUUGGCAUUUUGCCAAGAGCCC_CCAAUUGCAUACAUGUGCAUAAAUU .............................................((((((.....))))))...........(((((....)))))...... (-11.95 = -12.45 + 0.50)


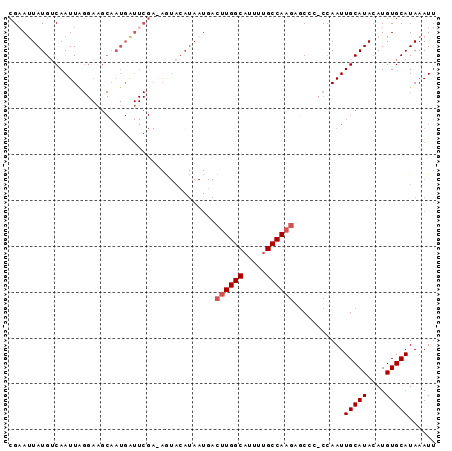
| Location | 3,108,094 – 3,108,186 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 74.46 |
| Mean single sequence MFE | -21.07 |
| Consensus MFE | -12.45 |
| Energy contribution | -15.20 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3108094 92 - 22224390 AAUUUAUGCACAUUUAUGCAAUUGG-GGGCUCUUGGCAAAAUGCCAAGUCAUUAUGUACUUUCGAACCAUUGCAUCCCAAUUGACAUAAUUCG .............((((((((((((-((((.((((((.....))))))......((......)).......)).))))))))).))))).... ( -25.60) >DroSec_CAF1 67208 74 - 1 AAUUUAUGCACAUGUAUGCAAUUGG-UUGCUCUUGGCAAAAUGCCAAGUCAUUAUGUACU------------------AAUUGACAUAGUUCG ..............(((((((((((-(.((.((((((.....)))))).......)))))------------------))))).))))..... ( -18.70) >DroEre_CAF1 67753 78 - 1 AAUUUAUGCACAUGUAUGCAAUUUGGGGGCUCUUGGCGAAAUGCCA---------------UCGAAUCAUUGCUUCCUAAUUGUCAUAAUUCG ..............(((((((((.((((((((.((((.....))))---------------..))......)))))).))))).))))..... ( -18.60) >DroYak_CAF1 72103 91 - 1 AAUUUAUGCACAUGUAUGCAAUUGGG--GGUCUUGGCAAAAUGCCAAGUAAUUAUGUAUUUUCGAAUCAUUGCUUCCUAAUUGACAUAAUACG ..............((((((((((((--((.((((((.....)))))).......(((............))))))))))))).))))..... ( -21.40) >consensus AAUUUAUGCACAUGUAUGCAAUUGG_GGGCUCUUGGCAAAAUGCCAAGUCAUUAUGUACU_UCGAAUCAUUGCUUCCUAAUUGACAUAAUUCG ..............(((((((((((((.((.((((((.....)))))).......................)).))))))))).))))..... (-12.45 = -15.20 + 2.75)

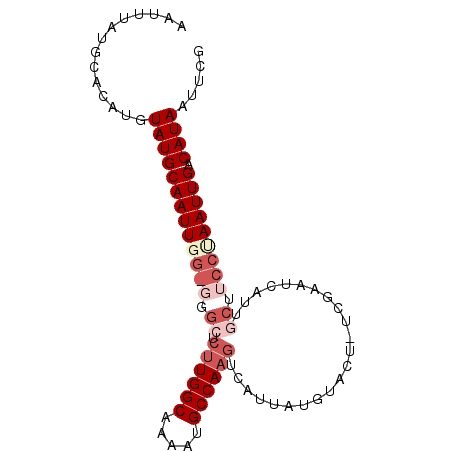
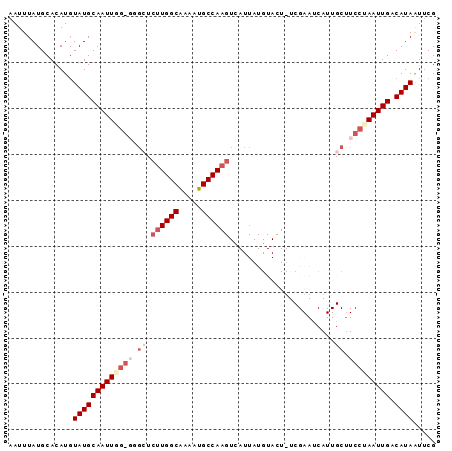
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:44 2006