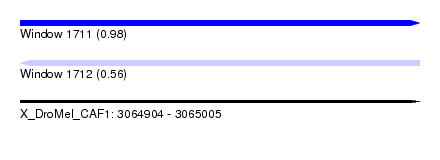
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,064,904 – 3,065,005 |
| Length | 101 |
| Max. P | 0.977086 |

| Location | 3,064,904 – 3,065,005 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -12.82 |
| Consensus MFE | -12.48 |
| Energy contribution | -11.47 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3064904 101 + 22224390 AAAAAGGUGGAAACAAAAAAAAUCUGGCAAAAUAAAGGAAAGCAAUGUAGCGAAGGCAGAAUGCUAAAAGGUAAAGUGCAAUCGAACAUUUUGUUAAUAUA .......((....)).........(((((((((....((..(((.....((....))....((((....))))...)))..))....)))))))))..... ( -16.40) >DroSec_CAF1 36486 96 + 1 AAAAAGGGAGAAAU-----AAAUCUGGCAAAAUAAAGGAAAGCAAUGUAGCAAAGGUAAAAUGCUAAAAGGUAAAGUGCAAUCGAACAUUUUGUUAAUAUA ..............-----.....(((((((((........((....(((((.........)))))....))...((........)))))))))))..... ( -11.10) >DroEre_CAF1 37272 95 + 1 AAAAAGA-A-----AAAAGAAAUCUGACGAAAUAAAGGAAAGCAAAGUAGCAAAGGCAGAAUGCUAAAAGGUAAAGUGCAAUCGAACAUUUUGUUAAUAUA .......-.-----..........(((((((((....((..(((...((.(...(((.....)))....).))...)))..))....)))))))))..... ( -12.80) >DroYak_CAF1 38133 89 + 1 AGAAAGA-A-----------AAUCUGGCGAAAUAAAGGAAAGCAAUGUAGCAAAGGCGGAAUGCUAAAAAGUAAAGUGCAAUCGAACAUUUUGUUAAUAUA .......-.-----------....(((((((((....((..(((.....((....))....((((....))))...)))..))....)))))))))..... ( -11.00) >consensus AAAAAGA_A__________AAAUCUGGCAAAAUAAAGGAAAGCAAUGUAGCAAAGGCAGAAUGCUAAAAGGUAAAGUGCAAUCGAACAUUUUGUUAAUAUA ........................(((((((((........((......))....(((...((((....))))...)))........)))))))))..... (-12.48 = -11.47 + -1.00)

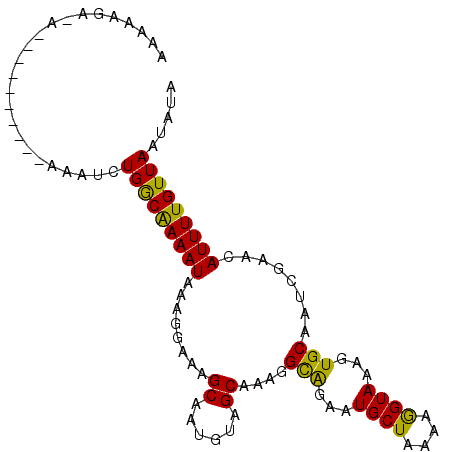
| Location | 3,064,904 – 3,065,005 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -8.58 |
| Consensus MFE | -7.02 |
| Energy contribution | -7.77 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3064904 101 - 22224390 UAUAUUAACAAAAUGUUCGAUUGCACUUUACCUUUUAGCAUUCUGCCUUCGCUACAUUGCUUUCCUUUAUUUUGCCAGAUUUUUUUUGUUUCCACCUUUUU ......(((((((...((....(((...........((((....((....)).....))))...........)))..))....)))))))........... ( -11.05) >DroSec_CAF1 36486 96 - 1 UAUAUUAACAAAAUGUUCGAUUGCACUUUACCUUUUAGCAUUUUACCUUUGCUACAUUGCUUUCCUUUAUUUUGCCAGAUUU-----AUUUCUCCCUUUUU ........(((((((...((..(((..........(((((.........)))))...)))..))...)))))))..(((...-----...)))........ ( -10.12) >DroEre_CAF1 37272 95 - 1 UAUAUUAACAAAAUGUUCGAUUGCACUUUACCUUUUAGCAUUCUGCCUUUGCUACUUUGCUUUCCUUUAUUUCGUCAGAUUUCUUUU-----U-UCUUUUU .......((.(((((...((..(((..........(((((.........)))))...)))..))...))))).))............-----.-....... ( -7.12) >DroYak_CAF1 38133 89 - 1 UAUAUUAACAAAAUGUUCGAUUGCACUUUACUUUUUAGCAUUCCGCCUUUGCUACAUUGCUUUCCUUUAUUUCGCCAGAUU-----------U-UCUUUCU ..........(((((...((..(((..........(((((.........)))))...)))..))...))))).........-----------.-....... ( -6.02) >consensus UAUAUUAACAAAAUGUUCGAUUGCACUUUACCUUUUAGCAUUCUGCCUUUGCUACAUUGCUUUCCUUUAUUUCGCCAGAUUU__________U_CCUUUUU ........(((((((...((..(((..........(((((.........)))))...)))..))...)))))))........................... ( -7.02 = -7.77 + 0.75)

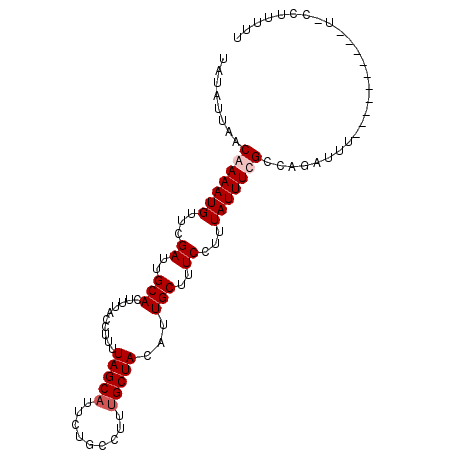
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:28 2006