| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,057,572 – 3,057,675 |
| Length | 103 |
| Max. P | 0.987475 |

| Location | 3,057,572 – 3,057,675 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.95 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -16.22 |
| Energy contribution | -17.38 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3057572 103 + 22224390 AAGUUUAAGGGCAGAUAUCCAAAUGCUUUUCUUGGCCAUAUGCAGA-GGAGCAUAAAAGCCAGAUAAAAAGAGGCUAUUAGGCUUAGGCCAAGAA------AAUUGUAGA ........(((......)))...((((((((((((((.(((((...-...))))).(((((.((((.........)))).))))).)))))))))------))..))).. ( -31.20) >DroPse_CAF1 27484 98 + 1 ---UGUGGUGGUAGCUGG---------CUUCUUGGCCAUAUGCAUACCGAGCAUAAAAGCCGGAUAAAAAGACGCUAUUAGGAUUAGUCAAAGAAGCAACCAAUAAGAGA ---.....((((.....(---------(((((((((..(((((.......)))))...))).........(((.((....))....))).))))))).))))........ ( -23.50) >DroSec_CAF1 29351 103 + 1 AAGUUUAAGGGCAGAUAUCCAAAUGCUUUUCUUGGCCAUAUGCAGA-AGAGCAUAAAAGCCAGAUAAGAAGAGGCUAUUAGGCUUAGGCCAAGAA------AAUGGCAGA ........(((......)))...((((((((((((((.(((((...-...))))).(((((.((((.(......))))).))))).)))))))))------)..)))).. ( -34.00) >DroEre_CAF1 29977 103 + 1 AAGUUUAAGGGCAGAUAUCCAAAUGCUUUUCUUGGCCAUAUGCAGA-GGAGCAUAAAAGCCAGAUAAAAAGAGGCUAUUAGGCUUAGGCCAAGAA------AAUGGCAGA ........(((......)))...((((((((((((((.(((((...-...))))).(((((.((((.........)))).))))).)))))))))------)..)))).. ( -33.90) >DroYak_CAF1 30912 103 + 1 AAGUUUAAGGGCAGAUAUCCAAAUGCUUUUCUUGGCCAUAUGCAGA-GGAGCAUAAAAUCCAGAUAAAAAGAGGCUAUUAGGCUUAGGCCAAGAA------AAUGGCAGA ........(((......)))...((((((((((((((.(((((...-...)))))................(((((....))))).)))))))))------)..)))).. ( -30.70) >DroPer_CAF1 27831 98 + 1 ---UGUGGUGGUAGCUAG---------CUUCUUGGCCAUAUGCAUACCGAGCAUAAAAGCCGGAUAAAAAGACGCUAUUAGGAUUAGUCAAAGAAGCAACCAAUAAGAGA ---.....((((.....(---------(((((((((..(((((.......)))))...))).........(((.((....))....))).))))))).))))........ ( -22.40) >consensus AAGUUUAAGGGCAGAUAUCCAAAUGCUUUUCUUGGCCAUAUGCAGA_GGAGCAUAAAAGCCAGAUAAAAAGAGGCUAUUAGGCUUAGGCCAAGAA______AAUGGCAGA ............................(((((((((.(((((.......)))))..((((...........))))..........)))))))))............... (-16.22 = -17.38 + 1.17)

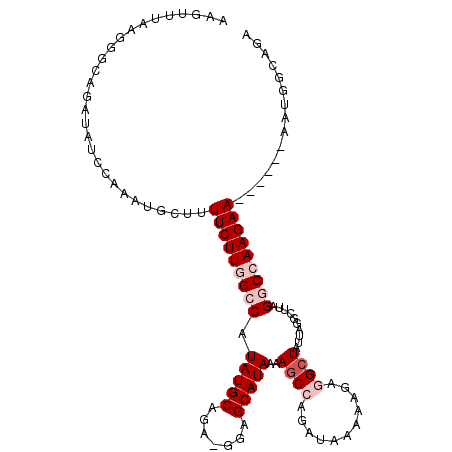
| Location | 3,057,572 – 3,057,675 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.95 |
| Mean single sequence MFE | -27.49 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.92 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3057572 103 - 22224390 UCUACAAUU------UUCUUGGCCUAAGCCUAAUAGCCUCUUUUUAUCUGGCUUUUAUGCUCC-UCUGCAUAUGGCCAAGAAAAGCAUUUGGAUAUCUGCCCUUAAACUU ((((...((------(((((((((.(((((..((((.......))))..))))).(((((...-...))))).))))))))))).....))))................. ( -29.70) >DroPse_CAF1 27484 98 - 1 UCUCUUAUUGGUUGCUUCUUUGACUAAUCCUAAUAGCGUCUUUUUAUCCGGCUUUUAUGCUCGGUAUGCAUAUGGCCAAGAAG---------CCAGCUACCACCACA--- ........((((((((((((.(((.............))).........((((..(((((.......))))).))))))))))---------.))))))........--- ( -24.32) >DroSec_CAF1 29351 103 - 1 UCUGCCAUU------UUCUUGGCCUAAGCCUAAUAGCCUCUUCUUAUCUGGCUUUUAUGCUCU-UCUGCAUAUGGCCAAGAAAAGCAUUUGGAUAUCUGCCCUUAAACUU ..(((..((------(((((((((.(((((..((((.......))))..))))).(((((...-...))))).))))))))))))))...((........))........ ( -30.50) >DroEre_CAF1 29977 103 - 1 UCUGCCAUU------UUCUUGGCCUAAGCCUAAUAGCCUCUUUUUAUCUGGCUUUUAUGCUCC-UCUGCAUAUGGCCAAGAAAAGCAUUUGGAUAUCUGCCCUUAAACUU ..(((..((------(((((((((.(((((..((((.......))))..))))).(((((...-...))))).))))))))))))))...((........))........ ( -30.50) >DroYak_CAF1 30912 103 - 1 UCUGCCAUU------UUCUUGGCCUAAGCCUAAUAGCCUCUUUUUAUCUGGAUUUUAUGCUCC-UCUGCAUAUGGCCAAGAAAAGCAUUUGGAUAUCUGCCCUUAAACUU ..(((..((------(((((((((....((..((((.......))))..))....(((((...-...))))).))))))))))))))...((........))........ ( -25.60) >DroPer_CAF1 27831 98 - 1 UCUCUUAUUGGUUGCUUCUUUGACUAAUCCUAAUAGCGUCUUUUUAUCCGGCUUUUAUGCUCGGUAUGCAUAUGGCCAAGAAG---------CUAGCUACCACCACA--- ........((((((((((((.(((.............))).........((((..(((((.......))))).))))))))))---------).)))))........--- ( -24.32) >consensus UCUGCCAUU______UUCUUGGCCUAAGCCUAAUAGCCUCUUUUUAUCUGGCUUUUAUGCUCC_UCUGCAUAUGGCCAAGAAAAGCAUUUGGAUAUCUGCCCUUAAACUU ...............(((((((((..........((((...........))))..(((((.......))))).)))))))))............................ (-15.75 = -16.92 + 1.17)
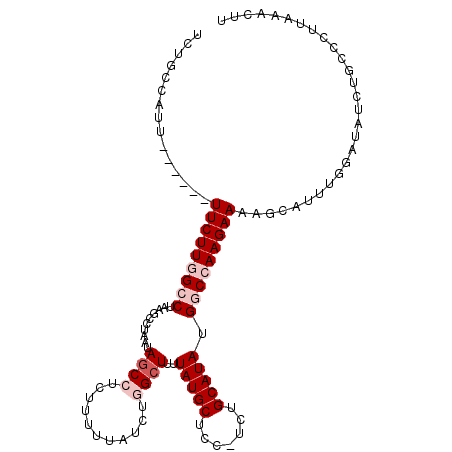
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:23 2006