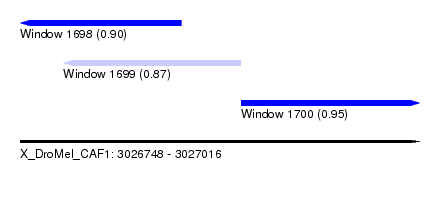
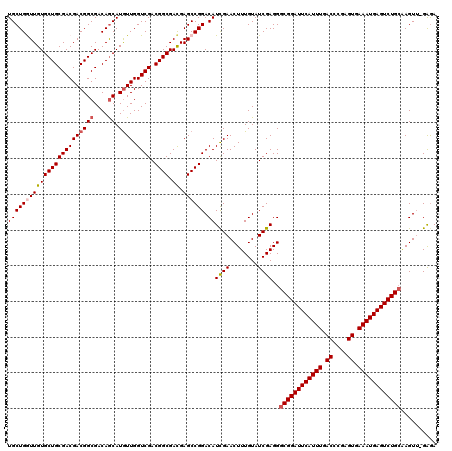
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,026,748 – 3,027,016 |
| Length | 268 |
| Max. P | 0.949234 |

| Location | 3,026,748 – 3,026,856 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.95 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -29.08 |
| Energy contribution | -29.36 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3026748 108 - 22224390 ACGGCGAUGAGCCGGACAUUGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUU-GGGAUGGUAGAAUGAUUAGUUAGCUAGU-----------UAGCU .((((.....))))..((((...(((((...)))))((((((((((((.((....)).)))))))))))).....-..))))......(((((((....)))))-----------))... ( -34.60) >DroSec_CAF1 7131 108 - 1 ACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUU-GGGAUGGUAGAACGAUUAGUCAGCUAGU-----------GAUAG .((((.....))))(((((((..(((..(..(((..((((((((((((.((....)).))))))))))))...))-)..)..).))..))))..)))..(((..-----------..))) ( -35.00) >DroSim_CAF1 2102 108 - 1 ACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUU-GAGAUGGUAGAACGAUUAGUCAGCUAGU-----------GAUAG .((((.....))))(((((((..(((..(.((((..((((((((((((.((....)).))))))))))))...))-)).)..).))..))))..)))..(((..-----------..))) ( -35.80) >DroEre_CAF1 7145 101 - 1 ACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGAGAGUGGGAAAUGUUGAAAUUAUUAGA-------------------GAUCA .((((.....))))((((((((........))))...(((((((((((.((....)).)))))))))))...........))))............-------------------..... ( -26.60) >DroYak_CAF1 7102 119 - 1 ACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUG-GAGAUGGUAAAGUUAUUAGUCAGCUUGCAUGGAAAUACGUAGCA .((((.....))))(((....((((((((.((.(..((((((((((((.((....)).))))))))))))...).-))....))))))))....))).(((((.((....)).)).))). ( -37.90) >consensus ACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUU_GAGAUGGUAGAAUGAUUAGUCAGCUAGU___________GAUCA .((((.....))))..((((...(((((...)))))((((((((((((.((....)).))))))))))))........))))...................................... (-29.08 = -29.36 + 0.28)

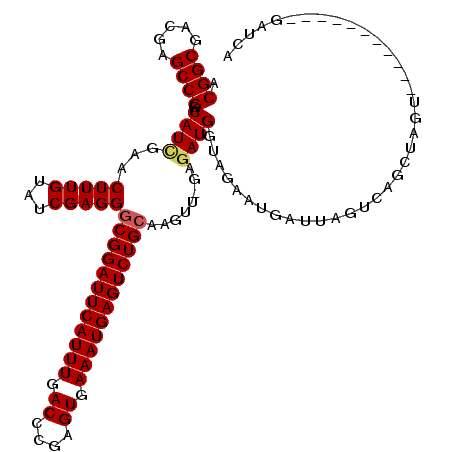
| Location | 3,026,777 – 3,026,896 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.81 |
| Mean single sequence MFE | -43.56 |
| Consensus MFE | -38.94 |
| Energy contribution | -39.62 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3026777 119 - 22224390 UGCUGAUUGUGCUGCGACGACGGCGACAACAUAUUGGUCGACGGCGAUGAGCCGGACAUUGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUU-GGGA .((((.((((......))))))))..((((.......((((((((.....)))).(((........))).))))..((((((((((((.((....)).))))))))))))..)))-)... ( -41.20) >DroSec_CAF1 7160 119 - 1 UGCUGGUUGUGCUGCGACGACGGCGACAGCAUGUUGGUCGACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUU-GGGA .(((.(((..((((((((((((((....)).)))).)))).))))))).)))(.(((.((((........))))..((((((((((((.((....)).))))))))))))..)))-.).. ( -46.10) >DroSim_CAF1 2131 119 - 1 UGCUGGUUGUGCUGCGACGACGGCGACAGCAUGUUGGUCGACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUU-GAGA ((((((((((((((((((((((((....)).)))).)))).)))).)).)))))).))((((.(((((...)))))((((((((((((.((....)).))))))))))))...))-)).. ( -46.30) >DroEre_CAF1 7166 120 - 1 UGCUGAUUGUGCUGCGACGACGGCGACAGCAUGUUGGUCGACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGAGAGUGGGAAA (((((.((((((((((((((((((....)).)))).)))).)))).))))..))).))((((........))))...(((((((((((.((....)).)))))))))))........... ( -38.50) >DroYak_CAF1 7142 119 - 1 UGCUGGUUGUGCUGCGACGACGGCGACAGCAUGUUGGUCGACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUG-GAGA ((((((((((((((((((((((((....)).)))).)))).)))).)).)))))).))((((........))))..((((((((((((.((....)).)))))))))))).....-.... ( -45.70) >consensus UGCUGGUUGUGCUGCGACGACGGCGACAGCAUGUUGGUCGACGGCGACGAGCCGGACAUCGAACUUUGUAUCGAGGGCGGAUUCAUUUGACCCGAGUGAAAUGAGUCUGCAAGUU_GAGA ((((((((((((((((((((((((....)).)))).)))).)))).)).)))))).))((((........))))..((((((((((((.((....)).)))))))))))).......... (-38.94 = -39.62 + 0.68)

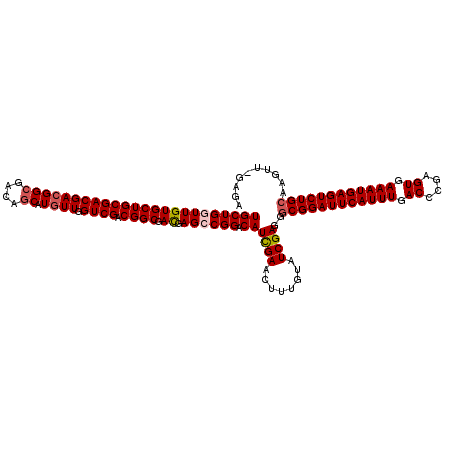
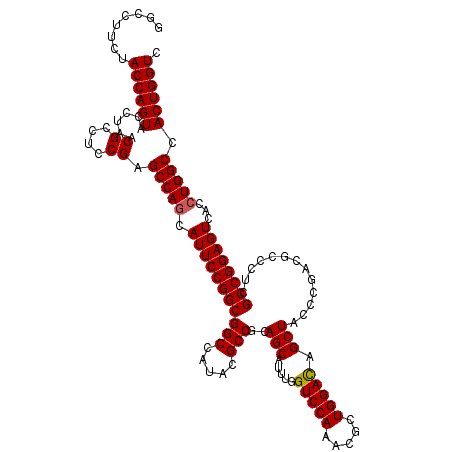
| Location | 3,026,896 – 3,027,016 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -40.38 |
| Energy contribution | -40.42 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3026896 120 + 22224390 GGCCUUCUACCAGUACCUAACGCCUUCGAGCCAACAUUCCGGCGGCCAUACGCCGCAGCAUUUGGUCCAAACGUUGGAUAGCUAUCCGACGCCCUCGCCGGAGUCACCUGGCCACUGGUC ........((((((..(....).....(.((((..(((((((((((((...((....))...)))))....((((((((....)))))))).....))))))))....))))))))))). ( -43.40) >DroSec_CAF1 7279 120 + 1 GGCCUUCUACCAGUACCUAACGCCUCCGAGCCAGCAUUCCGGCGGCCAUACGCCGCAGCACUUGGUCCAAACGCUGGACAGCUACCCGACGCCCUCGCCGGAGUCACCUGGCCACUGGUC ........((((((......((....)).(((((.(((((((((((.....)))..(((.....(((((.....))))).))).............))))))))...))))).)))))). ( -41.40) >DroSim_CAF1 2250 120 + 1 GGCCUUCUACCAGUACCUAACGCCUCCGAGCCAGCAUUCCGGCGGCCAUACGCCGCAGCACUUGGUCCAAACGCUGGACAGCUACCCGACGCCCUCGCCGGAGUCACCUGGCCACUGGUC ........((((((......((....)).(((((.(((((((((((.....)))..(((.....(((((.....))))).))).............))))))))...))))).)))))). ( -41.40) >DroEre_CAF1 7286 120 + 1 GGCCUUCUACCAGUACCUAACGCCUCCGAGCCAGCAUUCCGGCGGCCAUACGCCGCAGCAUUUGGUCCAAACGCUGGACAGCUAUCCAACGCCCUCGCCGGAGUCACCUGGCCACUGGUC ........((((((......((....)).(((((.(((((((((((.....)))..(((.....(((((.....))))).))).............))))))))...))))).)))))). ( -41.40) >DroYak_CAF1 7261 120 + 1 GGCCUUCUACCAGUACCUAACGCCACCGAGCCAGCAUUCCGGCGGCCAUACGCCGCAGCAUUUGGUCCAAACGCUGGACAGCUACCCAACGCCCUCGCCGGAGUCACCUGGCCACUGGUC ........((((((......((....)).(((((.(((((((((((.....)))..(((.....(((((.....))))).))).............))))))))...))))).)))))). ( -41.40) >consensus GGCCUUCUACCAGUACCUAACGCCUCCGAGCCAGCAUUCCGGCGGCCAUACGCCGCAGCAUUUGGUCCAAACGCUGGACAGCUACCCGACGCCCUCGCCGGAGUCACCUGGCCACUGGUC ........((((((......((....)).(((((.(((((((((((.....)))..(((.....(((((.....))))).))).............))))))))...))))).)))))). (-40.38 = -40.42 + 0.04)

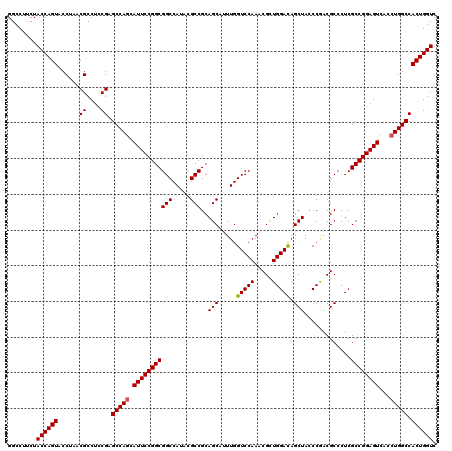
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:17 2006