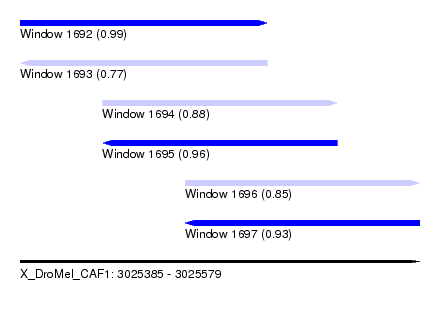
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,025,385 – 3,025,579 |
| Length | 194 |
| Max. P | 0.985607 |

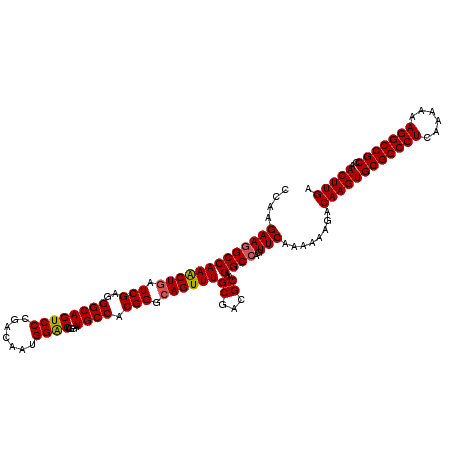
| Location | 3,025,385 – 3,025,505 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -44.77 |
| Consensus MFE | -42.31 |
| Energy contribution | -42.53 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3025385 120 + 22224390 CCAAGAAGGCCAAACUAAUCGAGGGCAGUCCGGACAAUGGGCUGGAUGCCACGGGCAGUUUGCGACGCAAGGCAAGUUCAAAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGA ....(((.((((((((..(((..((((.(((((........))))))))).)))..)))))((...))..)))...)))........((((((((((((......)))))))..))))). ( -43.50) >DroEre_CAF1 5822 120 + 1 CCAAGAAGGCCAAACUGAUCGAGGGCAGUCCCGACAAUGGACUGGAUGCCACGGGCAGUUUGCGACGCAAGGCCAGUUCAAAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGA .......((((((((((..((.((.((((((.......))))))....)).))..))))))((...))..)))).............((((((((((((......)))))))..))))). ( -45.50) >DroYak_CAF1 5750 120 + 1 CCAAGAAGGCCAAGCUGAUCGAGGGCAGUCCCGACAAUGGACUCGAUGCCACGGGCAGUUUGCGACGCAAGGCCAGUUCAAAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGA .......((((((((((..((..((((((((.......))))....)))).))..))))))((...))..)))).............((((((((((((......)))))))..))))). ( -45.30) >consensus CCAAGAAGGCCAAACUGAUCGAGGGCAGUCCCGACAAUGGACUGGAUGCCACGGGCAGUUUGCGACGCAAGGCCAGUUCAAAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGA ....(((((((((((((.(((..((((((((.......))))....)))).))).))))))((...))..))))..)))........((((((((((((......)))))))..))))). (-42.31 = -42.53 + 0.23)

| Location | 3,025,385 – 3,025,505 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -38.99 |
| Energy contribution | -39.77 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3025385 120 - 22224390 UCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUUUGAACUUGCCUUGCGUCGCAAACUGCCCGUGGCAUCCAGCCCAUUGUCCGGACUGCCCUCGAUUAGUUUGGCCUUCUUGG .((((..(((((((((....))))))(((....((......))...))).........(((((((..((.(((((((.((.....))..))).))))..))..))))))))))..)))). ( -39.90) >DroEre_CAF1 5822 120 - 1 UCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUUUGAACUGGCCUUGCGUCGCAAACUGCCCGUGGCAUCCAGUCCAUUGUCGGGACUGCCCUCGAUCAGUUUGGCCUUCUUGG .((((..(((((((((....))))))(((...(((...........)))..)))....(((((((..((.((....((((((.......)))))).)).))..))))))))))..)))). ( -44.90) >DroYak_CAF1 5750 120 - 1 UCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUUUGAACUGGCCUUGCGUCGCAAACUGCCCGUGGCAUCGAGUCCAUUGUCGGGACUGCCCUCGAUCAGCUUGGCCUUCUUGG .(((((..((((((((....))))))))))))).............((((..(((((((.........)))))(((((((((.......)))).....)))))..))..))))....... ( -43.50) >consensus UCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUUUGAACUGGCCUUGCGUCGCAAACUGCCCGUGGCAUCCAGUCCAUUGUCGGGACUGCCCUCGAUCAGUUUGGCCUUCUUGG .(((((..((((((((....)))))))))))))........(((..((((..(((((((.........)))))...((((((.......))))))..........))..))))))).... (-38.99 = -39.77 + 0.78)

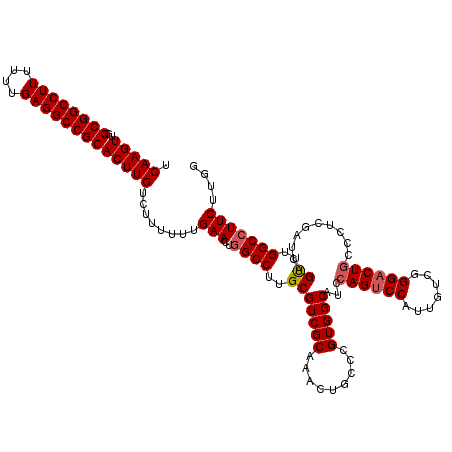
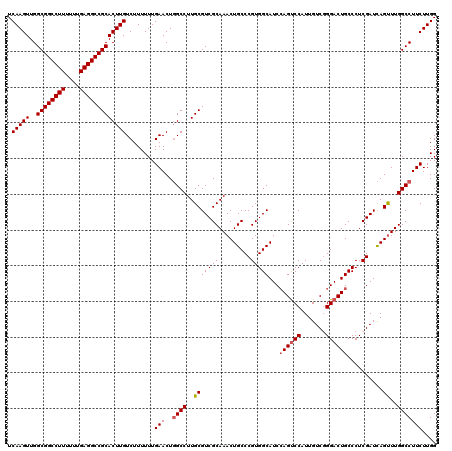
| Location | 3,025,425 – 3,025,539 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.43 |
| Mean single sequence MFE | -43.58 |
| Consensus MFE | -33.04 |
| Energy contribution | -33.20 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3025425 114 + 22224390 GCUGGAUGCCACGGGCAGUUUGCGACGCAAGGCAAGUUCAAAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGAAUG---GCCUAAAUCCGGGCCAGCUGACGGGGGG---AGU (((...(.((.((..((((((((..(....)))))............((((((((((((......)))))))..)))))..((---((((......)))))))))).)))).).---))) ( -40.20) >DroSec_CAF1 5844 111 + 1 ACUGGACGCCACGGGCAGCUUGCGACGCAAGGCCAGUUCAAAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACCUGAAUG---GCCUGAAUCCGGGCCAGCUGACGG---G---AGU (((((.((((...)))..(((((...))))))))))).............(.(((((((......))))))))..((((..((---(((((....))))))).....)))---)---... ( -44.00) >DroSim_CAF1 825 111 + 1 ACUGGACGCCACGGGCAGCUUGCGACGCAAGGCCAGUUCAAAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACCUGAAUG---GCCUGAAUCCGGGCCAGCUGACGG---G---CGU .....(((((.((..(((((.((..((..(((((((((.......)))..(.(((((((......))))))))........))---)))).....)).)).))))).)))---)---))) ( -47.00) >DroEre_CAF1 5862 116 + 1 ACUGGAUGCCACGGGCAGUUUGCGACGCAAGGCCAGUUCAAAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGAAUGGGGGCCUCAAUCCGGGCCAGCUGACGGGUGGAG---- .(((((((((...)))...((((...))))((((...((......))((((((((((((......)))))))..)))))......))))...)))))).(((.(.....).)))..---- ( -40.90) >DroYak_CAF1 5790 117 + 1 ACUCGAUGCCACGGGCAGUUUGCGACGCAAGGCCAGUUCAAAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGAAUG---GCCUGAAUCCGGGCCAGCUGACGGGGGGAGGAGU ((((..(.((.(((((...((((...)))).)))...(((.......((((((((((((......)))))))..)))))..((---(((((....)))))))..)))))..)).).)))) ( -45.80) >consensus ACUGGAUGCCACGGGCAGUUUGCGACGCAAGGCCAGUUCAAAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGAAUG___GCCUGAAUCCGGGCCAGCUGACGGG_GG___AGU ...........((..(((((.((..((..((((....((......))((((((((((((......)))))))..))))).......)))).....)).)).))))).))........... (-33.04 = -33.20 + 0.16)

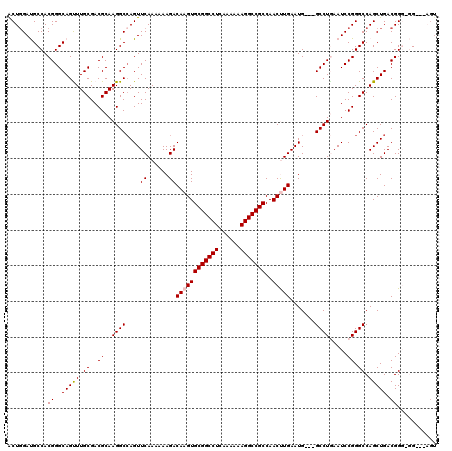
| Location | 3,025,425 – 3,025,539 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.43 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -35.38 |
| Energy contribution | -35.42 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3025425 114 - 22224390 ACU---CCCCCCGUCAGCUGGCCCGGAUUUAGGC---CAUUCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUUUGAACUUGCCUUGCGUCGCAAACUGCCCGUGGCAUCCAGC ...---..........((((((((((....((((---....(((((..((((((((....)))))))))))))((......))....))))......((.....))))).)))...)))) ( -37.40) >DroSec_CAF1 5844 111 - 1 ACU---C---CCGUCAGCUGGCCCGGAUUCAGGC---CAUUCAGGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUUUGAACUGGCCUUGCGUCGCAAGCUGCCCGUGGCGUCCAGU ...---.---......((((((((((....((((---((..(((((..((((((((....)))))))))))))((......))..)))))).(((.(....).)))))).)))...)))) ( -44.10) >DroSim_CAF1 825 111 - 1 ACG---C---CCGUCAGCUGGCCCGGAUUCAGGC---CAUUCAGGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUUUGAACUGGCCUUGCGUCGCAAGCUGCCCGUGGCGUCCAGU (((---(---(((.(((((.((...(((.(((((---((..(((((..((((((((....)))))))))))))((......))..))))).)).))))).)))))..)).)))))..... ( -48.50) >DroEre_CAF1 5862 116 - 1 ----CUCCACCCGUCAGCUGGCCCGGAUUGAGGCCCCCAUUCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUUUGAACUGGCCUUGCGUCGCAAACUGCCCGUGGCAUCCAGU ----............((((((((((...((((((......(((((..((((((((....)))))))))))))((......))...)))))).....((.....))))).)))...)))) ( -41.30) >DroYak_CAF1 5790 117 - 1 ACUCCUCCCCCCGUCAGCUGGCCCGGAUUCAGGC---CAUUCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUUUGAACUGGCCUUGCGUCGCAAACUGCCCGUGGCAUCGAGU ((((............((.((((.((.((((((.---....(((((..((((((((....))))))))))))).....))))))))))))..))(((((.........)))))...)))) ( -42.30) >consensus ACU___CC_CCCGUCAGCUGGCCCGGAUUCAGGC___CAUUCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUUUGAACUGGCCUUGCGUCGCAAACUGCCCGUGGCAUCCAGU ................((.((((.((.((((((........(((((..((((((((....))))))))))))).....))))))))))))..))(((((.........)))))....... (-35.38 = -35.42 + 0.04)


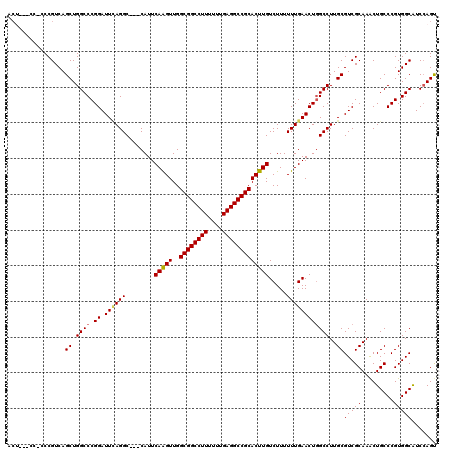
| Location | 3,025,465 – 3,025,579 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -48.58 |
| Consensus MFE | -33.37 |
| Energy contribution | -33.61 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3025465 114 + 22224390 AAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGAAUG---GCCUAAAUCCGGGCCAGCUGACGGGGGG---AGUGUCGGGCGUGCCGGGUGUACCGCCAACGAAUUCAGCGGCA .......((((((((((((......)))))))..)))))..((---((((......))))))(((((((((.((---.(..((.((....)).))..).)).))..))...))))).... ( -48.10) >DroSec_CAF1 5884 111 + 1 AAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACCUGAAUG---GCCUGAAUCCGGGCCAGCUGACGG---G---AGUGGCGGGCGUGCCGGGUGUACCGCCCUCGAAUUCAGCGGCA ..........(.(((((((......))))))))..((....((---(((((....)))))))(((((...---(---((.(((((..(..(...)..).))))))))....))))))).. ( -48.60) >DroSim_CAF1 865 111 + 1 AAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACCUGAAUG---GCCUGAAUCCGGGCCAGCUGACGG---G---CGUGUCGGGCGUGCCGGGUGUACCGCCCUCGAAUUCAGCGGCA ..........(.(((((((......))))))))..((....((---(((((....)))))))(((((..(---(---((((.....))))))(((((...)))))......))))))).. ( -47.60) >DroEre_CAF1 5902 108 + 1 AAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGAAUGGGGGCCUCAAUCCGGGCCAGCUGACGGGUGGAG------------UGCCGGGUGUACCGCCAACGAAUUCAGCGGCA .......((((((((((((......)))))))..)))))......(((((......))))).((((((((((((.(------------..(...)..).)))))..))...))))).... ( -41.80) >DroYak_CAF1 5830 117 + 1 AAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGAAUG---GCCUGAAUCCGGGCCAGCUGACGGGGGGAGGAGUGUCGGGUGCGCCGGGUGUACCGCCCUCGAAUUCGGCGGCA .......((((((((((((......)))))))..)))))..((---(((((....)))))))(((((((((((..((.(..((.((....)).))..).)).))))))...))))).... ( -56.80) >consensus AAAAAGACAAGUGCGGCCUCAAAAAAGGCCGCCAACUUGAAUG___GCCUGAAUCCGGGCCAGCUGACGGG_GG___AGUGUCGGGCGUGCCGGGUGUACCGCCCUCGAAUUCAGCGGCA .......((((((((((((......)))))))..)))))........((.......))(((.((((((((....................)))((((...)))).......)))))))). (-33.37 = -33.61 + 0.24)

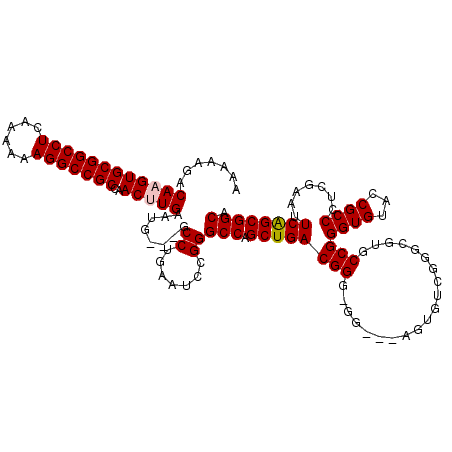
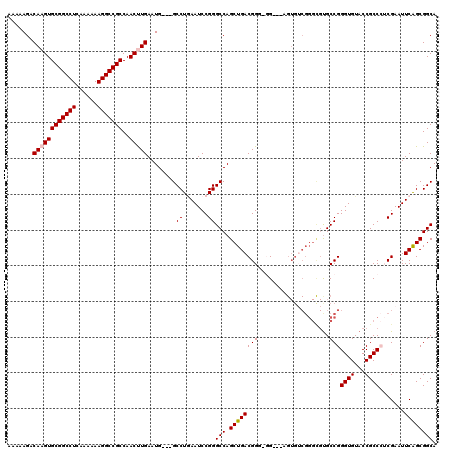
| Location | 3,025,465 – 3,025,579 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -43.94 |
| Consensus MFE | -37.55 |
| Energy contribution | -37.39 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3025465 114 - 22224390 UGCCGCUGAAUUCGUUGGCGGUACACCCGGCACGCCCGACACU---CCCCCCGUCAGCUGGCCCGGAUUUAGGC---CAUUCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUU ((((((..(.....)..)))))).....((....)).((((..---........(((((((((........)))---).....)))))((((((((....))))))))...))))..... ( -41.50) >DroSec_CAF1 5884 111 - 1 UGCCGCUGAAUUCGAGGGCGGUACACCCGGCACGCCCGCCACU---C---CCGUCAGCUGGCCCGGAUUCAGGC---CAUUCAGGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUU .(((..((((((((.(((((((.......)).)))))((((((---.---.....)).)))).)))))))))))---....(((((..((((((((....)))))))))))))....... ( -47.60) >DroSim_CAF1 865 111 - 1 UGCCGCUGAAUUCGAGGGCGGUACACCCGGCACGCCCGACACG---C---CCGUCAGCUGGCCCGGAUUCAGGC---CAUUCAGGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUU .(((..((((((((.(((((((.......)).))))).....(---(---(.(....).))).)))))))))))---....(((((..((((((((....)))))))))))))....... ( -46.50) >DroEre_CAF1 5902 108 - 1 UGCCGCUGAAUUCGUUGGCGGUACACCCGGCA------------CUCCACCCGUCAGCUGGCCCGGAUUGAGGCCCCCAUUCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUU ((((((..(.....)..)))))).....(((.------------(((..((.(((....)))..))...))))))......(((((..((((((((....)))))))))))))....... ( -40.40) >DroYak_CAF1 5830 117 - 1 UGCCGCCGAAUUCGAGGGCGGUACACCCGGCGCACCCGACACUCCUCCCCCCGUCAGCUGGCCCGGAUUCAGGC---CAUUCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUU ....((((((((((.((.((((......((.....))(((............))).)))).))))))))).)))---....(((((..((((((((....)))))))))))))....... ( -43.70) >consensus UGCCGCUGAAUUCGAGGGCGGUACACCCGGCACGCCCGACACU___CC_CCCGUCAGCUGGCCCGGAUUCAGGC___CAUUCAAGUUGGCGGCCUUUUUUGAGGCCGCACUUGUCUUUUU .(((...(((((((..(((((....))((((.........................)))))))))))))).))).......(((((..((((((((....)))))))))))))....... (-37.55 = -37.39 + -0.16)

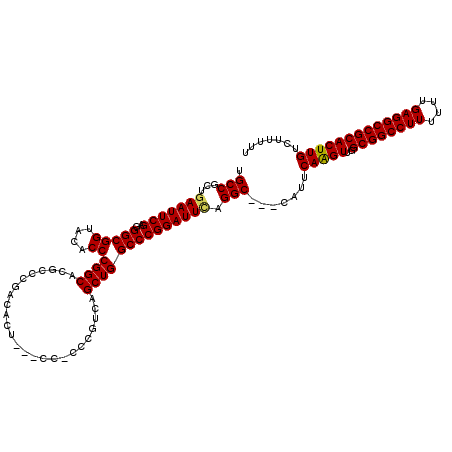
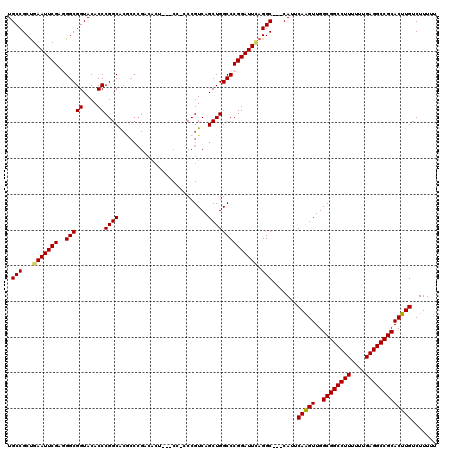
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:14 2006