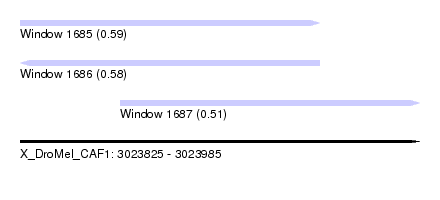
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,023,825 – 3,023,985 |
| Length | 160 |
| Max. P | 0.593636 |

| Location | 3,023,825 – 3,023,945 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -39.72 |
| Consensus MFE | -35.85 |
| Energy contribution | -35.85 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
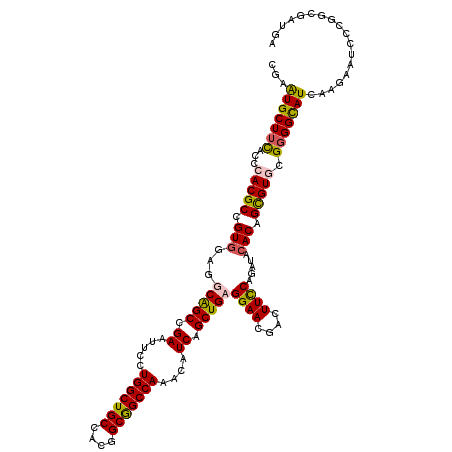
>X_DroMel_CAF1 3023825 120 + 22224390 GCAUUCAAAUCUAUUUGGAGAUUGACAAUCGCAAAUGCACCGAAUGCUUUACGCACGCCGUUGAGGCAGCCGAAUUUCUGGCUGCCACGGCGGCCAAACAUCAGCUGCGGAACGAUUUUC ................((((((((......((....)).(((...(((........(((((((.((((((((......)))))))).)))))))........)))..)))..)))))))) ( -42.89) >DroSec_CAF1 4244 120 + 1 GCAUUCAAAUCUAUUUGGAGAUUGACAAUCGCAAGUGCACCGAGUGCUUCACCCACGCCGUGGAGGCGGCCGAGUUCCUGGCUGCCACGGCGGCCAAACAUCAGCUGAGGAACGACUUCC ((((((.(((((......))))).......((....))...)))))).........(((((.(.((((((((......)))))))).).)))))............((((.....)))). ( -40.40) >DroEre_CAF1 4262 120 + 1 GCAUACAAAUCUAUCUGGAGAUUGAUAACCGCAAGUGCACCGAGUGCUUCACACACGCCGUGGAGGCAGCCGAAUUUCUGGCUGCCACGGCGGCCAAACAUCAGCUGAGGAACGACUUUC .............(((..((.(((((....(.(((..(.....)..))))......(((((.(.((((((((......)))))))).).))))).....)))))))..)))......... ( -38.70) >DroYak_CAF1 4190 120 + 1 GCAUACAAAUCUAUCUGGAGAUUGACAAUCGCAAGUGCACCGAAUGCUUCACCCACGCAGUGGAGGCGGCCGAAUUCCUGGCUGCCACGGCAGCCAAACAUCAGCUGCGGAAUGACUUCC (((....(((((......)))))......(....)))).((.....((((((.......))))))(((((.((.....(((((((....)))))))....)).))))))).......... ( -36.90) >consensus GCAUACAAAUCUAUCUGGAGAUUGACAAUCGCAAGUGCACCGAAUGCUUCACCCACGCCGUGGAGGCAGCCGAAUUCCUGGCUGCCACGGCGGCCAAACAUCAGCUGAGGAACGACUUCC ((((((.(((((......))))).......((....))...)))))).........(((((.(.((((((((......)))))))).).)))))..............((((....)))) (-35.85 = -35.85 + 0.00)

| Location | 3,023,825 – 3,023,945 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -38.21 |
| Energy contribution | -38.40 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3023825 120 - 22224390 GAAAAUCGUUCCGCAGCUGAUGUUUGGCCGCCGUGGCAGCCAGAAAUUCGGCUGCCUCAACGGCGUGCGUAAAGCAUUCGGUGCAUUUGCGAUUGUCAAUCUCCAAAUAGAUUUGAAUGC ((.((((((...(((.(.(((((((.(((((((((((((((........)))))))...)))))).))...))))))).).)))....)))))).))(((((......)))))....... ( -48.00) >DroSec_CAF1 4244 120 - 1 GGAAGUCGUUCCUCAGCUGAUGUUUGGCCGCCGUGGCAGCCAGGAACUCGGCCGCCUCCACGGCGUGGGUGAAGCACUCGGUGCACUUGCGAUUGUCAAUCUCCAAAUAGAUUUGAAUGC ...((((((......((((((((((.(((((((((((.(((........))).)))...)))))...))).))))).)))))......)))))).(((((((......))).)))).... ( -41.10) >DroEre_CAF1 4262 120 - 1 GAAAGUCGUUCCUCAGCUGAUGUUUGGCCGCCGUGGCAGCCAGAAAUUCGGCUGCCUCCACGGCGUGUGUGAAGCACUCGGUGCACUUGCGGUUAUCAAUCUCCAGAUAGAUUUGUAUGC ((.(..(((......((((((((((.(((((((((((((((........)))))))...))))))...)).))))).)))))......)))..).))(((((......)))))....... ( -43.10) >DroYak_CAF1 4190 120 - 1 GGAAGUCAUUCCGCAGCUGAUGUUUGGCUGCCGUGGCAGCCAGGAAUUCGGCCGCCUCCACUGCGUGGGUGAAGCAUUCGGUGCACUUGCGAUUGUCAAUCUCCAGAUAGAUUUGUAUGC ((((....))))(((.(.(((((((((((((....))))))))...((((.(((((......).)))).))))))))).).)))...(((((..(((.(((....))).))))))))... ( -42.90) >consensus GAAAGUCGUUCCGCAGCUGAUGUUUGGCCGCCGUGGCAGCCAGAAAUUCGGCCGCCUCCACGGCGUGGGUGAAGCACUCGGUGCACUUGCGAUUGUCAAUCUCCAAAUAGAUUUGAAUGC ((.((((((......((((((((((.(((((((((((((((........)))))))...))))))...)).))))).)))))......)))))).))(((((......)))))....... (-38.21 = -38.40 + 0.19)

| Location | 3,023,865 – 3,023,985 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -44.85 |
| Consensus MFE | -36.59 |
| Energy contribution | -36.52 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3023865 120 + 22224390 CGAAUGCUUUACGCACGCCGUUGAGGCAGCCGAAUUUCUGGCUGCCACGGCGGCCAAACAUCAGCUGCGGAACGAUUUUCAGAUACACAGUGUUAGGGGCAUCAAGAAUCCCGGCGAUGA ....(((.....))).(((((((.((((((((......)))))))).)))))))....((((.((((.(((..((((..(.(((((...))))).)..).))).....))))))))))). ( -47.00) >DroSec_CAF1 4284 120 + 1 CGAGUGCUUCACCCACGCCGUGGAGGCGGCCGAGUUCCUGGCUGCCACGGCGGCCAAACAUCAGCUGAGGAACGACUUCCAGAUACACAGUGUGCGGGGCAUCAAGAAUCCCGGCGAUGA ...(((.......)))(((((.(.((((((((......)))))))).).)))))....((((.((((.(((..((..(((.(.(((.....)))).)))..)).....))))))))))). ( -43.90) >DroEre_CAF1 4302 120 + 1 CGAGUGCUUCACACACGCCGUGGAGGCAGCCGAAUUUCUGGCUGCCACGGCGGCCAAACAUCAGCUGAGGAACGACUUUCAGAUACACAGCGUGCGGGGCAUCAAGAAUCCCGGCGAUGA ...(((.....)))..(((((.(.((((((((......)))))))).).)))))....((((.((((.(...(........)...).))))..((.(((.((.....))))).)))))). ( -45.90) >DroYak_CAF1 4230 120 + 1 CGAAUGCUUCACCCACGCAGUGGAGGCGGCCGAAUUCCUGGCUGCCACGGCAGCCAAACAUCAGCUGCGGAAUGACUUCCAGAUACACAGCGUUCGUGGUAUUAAGAAUCCCGGCGACGA ..(((((..(((..((((.(((...(((((.((.....(((((((....)))))))....)).)))))((((....)))).....))).))))..))))))))........((....)). ( -42.60) >consensus CGAAUGCUUCACCCACGCCGUGGAGGCAGCCGAAUUCCUGGCUGCCACGGCGGCCAAACAUCAGCUGAGGAACGACUUCCAGAUACACAGCGUGCGGGGCAUCAAGAAUCCCGGCGAUGA ...(((((((...(((((.(((...(((((.((.....(((((((....)))))))....)).)))))((((....)))).....))).))))).))))))).................. (-36.59 = -36.52 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:05 2006