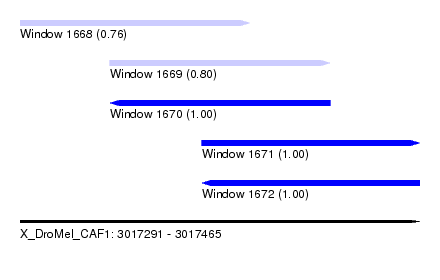
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,017,291 – 3,017,465 |
| Length | 174 |
| Max. P | 0.999969 |

| Location | 3,017,291 – 3,017,391 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.20 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -24.06 |
| Energy contribution | -24.26 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3017291 100 + 22224390 -GCUUUCUGGCUCAUUAGAAUUUUUGAUUGGUCGCAAUUAAAAGGCCAGACGACGAAUAUAUAGCGAAGCUUAUCAAAUUGUUUUAGCUACCCUCGCUGCA -((..((((((((....)).((((((((((....))))))))))))))))............(((((((((...(.....)....))))....))))))). ( -24.50) >DroSec_CAF1 5002 99 + 1 -GCUUUCUGGCUCAUUAGAAUUUUUGAUUGGUCGCAAUUAAAAGGCCAGACGAAG-AUAGAGAGCGAAGCUUAUCAAAUUGUUUUAGCUCCCCUCGCUGCA -((..((((((((....)).((((((((((....))))))))))))))))....(-(..(.(((((((((..........))))).)))))..))...)). ( -25.10) >DroSim_CAF1 5229 99 + 1 -GCUUUCUGGCUCAUUAGAAUUUUUGAUUGGUCGCAAUUAAAAGGCCAGACGAAG-AUACAGAGCGAAGCUUAUCAAAUUGUUUUUGCUCCCCUCGCUGCA -((..((((((((....)).((((((((((....)))))))))))))))).....-.....(((((((((..........).))))))))........)). ( -23.90) >DroEre_CAF1 5255 100 + 1 UGCUUUCUGGCUCAUUAGAAUUUUUGAUUGGACGCAAUUAAAAGGCCAGACGAAG-UUGCAGAGCGAAGCUUAUCAAAUUGUUUUAGCUCCCCUCGCUGCA .((((((((((((....)).((((((((((....))))))))))))))))..)))-)((((..((((((((...(.....)....))))....)))))))) ( -25.60) >DroYak_CAF1 5182 100 + 1 CGCUUUCUGGCUCAUUAGAAUUUUUGAUUGGACGCAAUUAAAAGGCCAGACGAAG-AUACAGAGCGAAGCUUAUCAAAUUGUUUUAGCUCCCCUCGCUGCA .((..((((((((....)).((((((((((....)))))))))))))))).....-......(((((((((...(.....)....))))....))))))). ( -25.00) >consensus _GCUUUCUGGCUCAUUAGAAUUUUUGAUUGGUCGCAAUUAAAAGGCCAGACGAAG_AUACAGAGCGAAGCUUAUCAAAUUGUUUUAGCUCCCCUCGCUGCA .((..((((((((....)).((((((((((....))))))))))))))))............(((((((((...(.....)....))))....))))))). (-24.06 = -24.26 + 0.20)

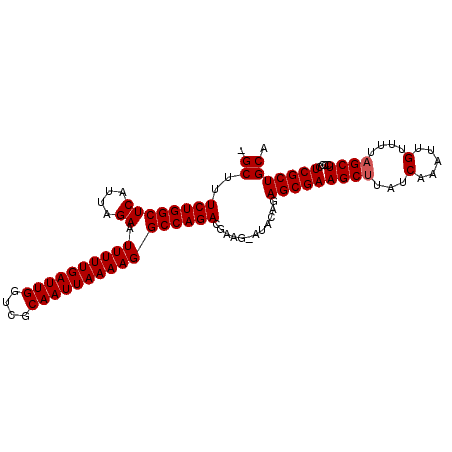
| Location | 3,017,330 – 3,017,426 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 95.70 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -24.54 |
| Energy contribution | -24.22 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3017330 96 + 22224390 AAAGGCCAGACGACGAAUAUAUAGCGAAGCUUAUCAAAUUGUUUUAGCUACCCUCGCUGCAGUUUGUGCAGCUUGUAAAUUGUCGUGUGCAAUUCG ....((.(.(((((((.....(((((((((..........))))).)))).....((((((.....)))))).......))))))).)))...... ( -26.40) >DroSec_CAF1 5041 95 + 1 AAAGGCCAGACGAAG-AUAGAGAGCGAAGCUUAUCAAAUUGUUUUAGCUCCCCUCGCUGCAGUUUGUGCAGCCUGUAAAUUGUCGUGUGCAAUUCG ....(((((((((..-((((.(((((((((..........))))).)))).....((((((.....))))))))))...))))).)).))...... ( -27.10) >DroSim_CAF1 5268 95 + 1 AAAGGCCAGACGAAG-AUACAGAGCGAAGCUUAUCAAAUUGUUUUUGCUCCCCUCGCUGCAGUUUGUGCAGCCUGUAAAUUGUCGUGUGCAAUUCG ....(((((((((..-.(((((((((((((..........).)))))))).....((((((.....)))))).))))..))))).)).))...... ( -26.10) >DroEre_CAF1 5295 95 + 1 AAAGGCCAGACGAAG-UUGCAGAGCGAAGCUUAUCAAAUUGUUUUAGCUCCCCUCGCUGCAGUUUGUGCAGCUUGUAAAUUGUCGUGUGCAAUUCG ....(((((((((..-((((((((((((((..........))))).)))).....((((((.....)))))).))))).))))).)).))...... ( -28.10) >DroYak_CAF1 5222 95 + 1 AAAGGCCAGACGAAG-AUACAGAGCGAAGCUUAUCAAAUUGUUUUAGCUCCCCUCGCUGCAGUUUGUGCAGCUUGUAAAUUGUCGUGUGCAAUUCG ....(((((((((..-.(((((((((((((..........))))).)))).....((((((.....)))))).))))..))))).)).))...... ( -26.60) >consensus AAAGGCCAGACGAAG_AUACAGAGCGAAGCUUAUCAAAUUGUUUUAGCUCCCCUCGCUGCAGUUUGUGCAGCUUGUAAAUUGUCGUGUGCAAUUCG ....(((((((((........(((((((((..........))))).)))).....((((((.....)))))).......))))).)).))...... (-24.54 = -24.22 + -0.32)

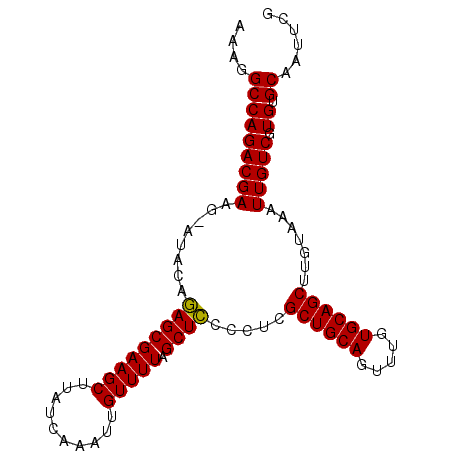
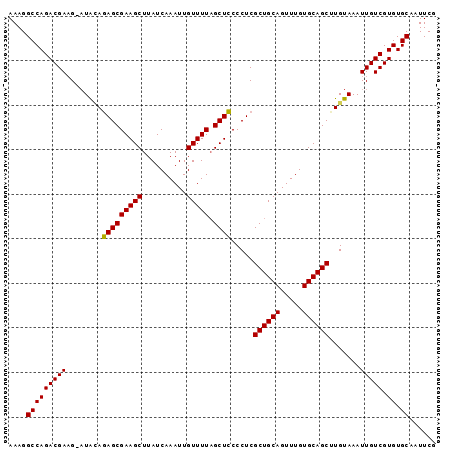
| Location | 3,017,330 – 3,017,426 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 95.70 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -24.80 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3017330 96 - 22224390 CGAAUUGCACACGACAAUUUACAAGCUGCACAAACUGCAGCGAGGGUAGCUAAAACAAUUUGAUAAGCUUCGCUAUAUAUUCGUCGUCUGGCCUUU ......((..(((((.........((((((.....))))))(((.(((((.....................)))))...))))))))...)).... ( -23.90) >DroSec_CAF1 5041 95 - 1 CGAAUUGCACACGACAAUUUACAGGCUGCACAAACUGCAGCGAGGGGAGCUAAAACAAUUUGAUAAGCUUCGCUCUCUAU-CUUCGUCUGGCCUUU ......((.((.(((.........((((((.....))))))(((.((((((..............)))))).))).....-....))))))).... ( -27.24) >DroSim_CAF1 5268 95 - 1 CGAAUUGCACACGACAAUUUACAGGCUGCACAAACUGCAGCGAGGGGAGCAAAAACAAUUUGAUAAGCUUCGCUCUGUAU-CUUCGUCUGGCCUUU ......((.((.(((((..((((.((((((.....))))))(((.(((((................))))).))))))).-.)).))))))).... ( -27.79) >DroEre_CAF1 5295 95 - 1 CGAAUUGCACACGACAAUUUACAAGCUGCACAAACUGCAGCGAGGGGAGCUAAAACAAUUUGAUAAGCUUCGCUCUGCAA-CUUCGUCUGGCCUUU (((((((((...............((((((.....))))))(((.((((((..............)))))).))))))))-.)))).......... ( -28.94) >DroYak_CAF1 5222 95 - 1 CGAAUUGCACACGACAAUUUACAAGCUGCACAAACUGCAGCGAGGGGAGCUAAAACAAUUUGAUAAGCUUCGCUCUGUAU-CUUCGUCUGGCCUUU ......((.((.(((((..((((.((((((.....))))))(((.((((((..............)))))).))))))).-.)).))))))).... ( -29.24) >consensus CGAAUUGCACACGACAAUUUACAAGCUGCACAAACUGCAGCGAGGGGAGCUAAAACAAUUUGAUAAGCUUCGCUCUGUAU_CUUCGUCUGGCCUUU ......((.((.(((.........((((((.....))))))(((.((((((..............)))))).)))..........))))))).... (-24.80 = -25.40 + 0.60)

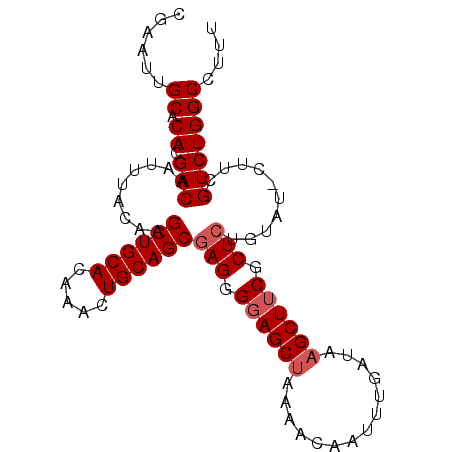
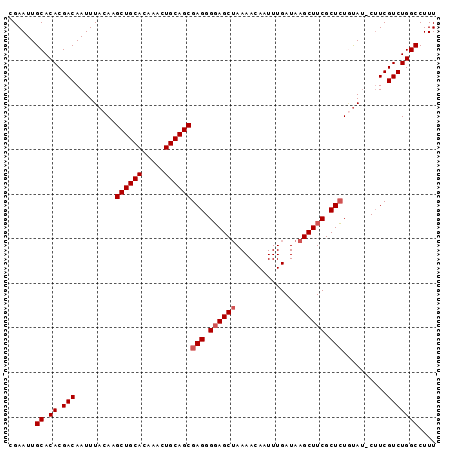
| Location | 3,017,370 – 3,017,465 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 97.07 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.03 |
| SVM RNA-class probability | 0.999969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3017370 95 + 22224390 GUUUUAGCUACCCUCGCUGCAGUUUGUGCAGCUUGUAAAUUGUCGUGUGCAAUUCGCAUCAAUUGGAAUGAAUUAAUUGAAAAUUGCACACAAA-C ...............((((((.....))))))............((((((((((....((((((((......)))))))).))))))))))...-. ( -26.80) >DroSec_CAF1 5080 95 + 1 GUUUUAGCUCCCCUCGCUGCAGUUUGUGCAGCCUGUAAAUUGUCGUGUGCAAUUCGCAUCAAUUGAAAUGAAUUAAUUGAAAAUUGCACACAAA-C ...............((((((.....))))))............((((((((((....((((((((......)))))))).))))))))))...-. ( -27.70) >DroSim_CAF1 5307 95 + 1 GUUUUUGCUCCCCUCGCUGCAGUUUGUGCAGCCUGUAAAUUGUCGUGUGCAAUUCGCAUCAAUUGAAAUGAAUUAAUUGAAAAUUGCACACAAA-C ...(((((.......((((((.....))))))..))))).....((((((((((....((((((((......)))))))).))))))))))...-. ( -28.00) >DroEre_CAF1 5334 96 + 1 GUUUUAGCUCCCCUCGCUGCAGUUUGUGCAGCUUGUAAAUUGUCGUGUGCAAUUCGCAUCAAUUGAAAUGAAUUAAUUGAAAAUUGCACACAAAAC (((((..........((((((.....))))))............((((((((((....((((((((......)))))))).))))))))))))))) ( -28.90) >DroYak_CAF1 5261 96 + 1 GUUUUAGCUCCCCUCGCUGCAGUUUGUGCAGCUUGUAAAUUGUCGUGUGCAAUUCGCAUCAAUUGAAAUGAAUUAAUUGAAAAUUGCACAAAAAAC (((((..........((((((.....)))))).............(((((((((....((((((((......)))))))).))))))))).))))) ( -25.20) >consensus GUUUUAGCUCCCCUCGCUGCAGUUUGUGCAGCUUGUAAAUUGUCGUGUGCAAUUCGCAUCAAUUGAAAUGAAUUAAUUGAAAAUUGCACACAAA_C ...............((((((.....))))))............((((((((((....((((((((......)))))))).))))))))))..... (-26.70 = -26.74 + 0.04)

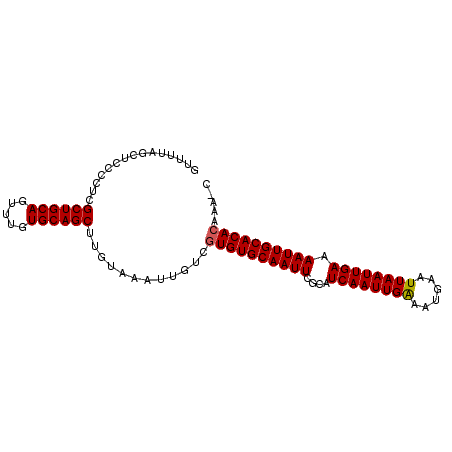
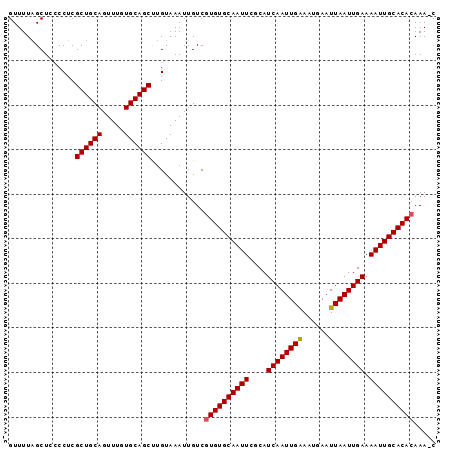
| Location | 3,017,370 – 3,017,465 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 97.07 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.40 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.87 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
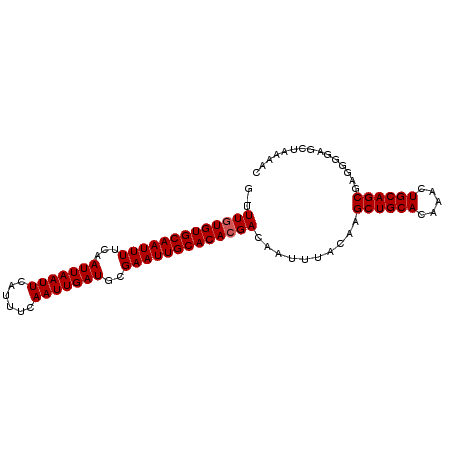
>X_DroMel_CAF1 3017370 95 - 22224390 G-UUUGUGUGCAAUUUUCAAUUAAUUCAUUCCAAUUGAUGCGAAUUGCACACGACAAUUUACAAGCUGCACAAACUGCAGCGAGGGUAGCUAAAAC .-.(((((((((((((...(((((((......)))))))..)))))))))))))....((((..((((((.....))))))....))))....... ( -30.80) >DroSec_CAF1 5080 95 - 1 G-UUUGUGUGCAAUUUUCAAUUAAUUCAUUUCAAUUGAUGCGAAUUGCACACGACAAUUUACAGGCUGCACAAACUGCAGCGAGGGGAGCUAAAAC (-((((((((((((((...(((((((......)))))))..)))))))))))............((((((.....)))))).....))))...... ( -29.40) >DroSim_CAF1 5307 95 - 1 G-UUUGUGUGCAAUUUUCAAUUAAUUCAUUUCAAUUGAUGCGAAUUGCACACGACAAUUUACAGGCUGCACAAACUGCAGCGAGGGGAGCAAAAAC (-((((((((((((((...(((((((......)))))))..)))))))))))............((((((.....)))))).....))))...... ( -29.90) >DroEre_CAF1 5334 96 - 1 GUUUUGUGUGCAAUUUUCAAUUAAUUCAUUUCAAUUGAUGCGAAUUGCACACGACAAUUUACAAGCUGCACAAACUGCAGCGAGGGGAGCUAAAAC ((((((((((((((((...(((((((......)))))))..)))))))))))............((((((.....))))))....)))))...... ( -30.10) >DroYak_CAF1 5261 96 - 1 GUUUUUUGUGCAAUUUUCAAUUAAUUCAUUUCAAUUGAUGCGAAUUGCACACGACAAUUUACAAGCUGCACAAACUGCAGCGAGGGGAGCUAAAAC ((((((((((((((((...(((((((......)))))))..)))))))))..............((((((.....))))))..)))))))...... ( -26.70) >consensus G_UUUGUGUGCAAUUUUCAAUUAAUUCAUUUCAAUUGAUGCGAAUUGCACACGACAAUUUACAAGCUGCACAAACUGCAGCGAGGGGAGCUAAAAC ...(((((((((((((...(((((((......)))))))..)))))))))))))..........((((((.....))))))............... (-28.20 = -28.40 + 0.20)

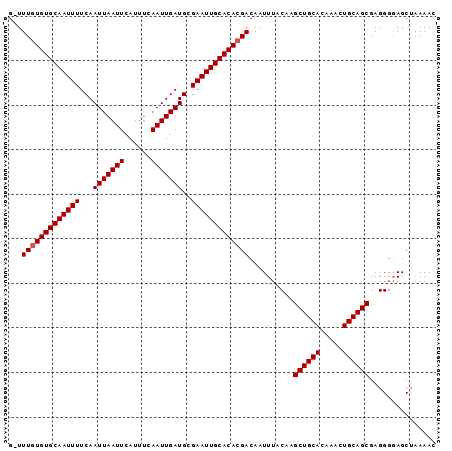
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:50 2006