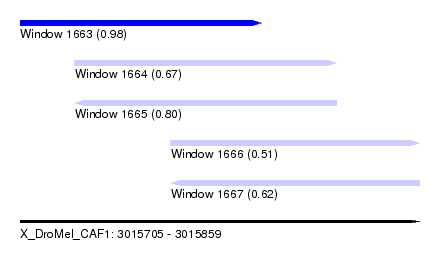
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,015,705 – 3,015,859 |
| Length | 154 |
| Max. P | 0.975043 |

| Location | 3,015,705 – 3,015,798 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 83.88 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -19.70 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3015705 93 + 22224390 GUUCUCAGCAAUCAGGCGAAUGUUUCGACGCUUUUCGAUGAGAACGUUUCUUCGUUCGCCUAUCGAUAACCAGUUAUCGAUGAUGGUGAGUCA ...((((.((.(((((((((((....((((.(((((...)))))))))....)))))))))((((((((....)))))))))))).))))... ( -34.40) >DroSec_CAF1 3520 89 + 1 GUUCUCAGCAAUCAGG--AACGUUUCGCUGCUUUCCGAUGAGAACGUUUCUUAG--CGCCUAUCGAUAACCAGUUAUCGAUGAUGGUAAGUCA (((((((.......((--((.((......)).))))..)))))))....((((.--((..(((((((((....))))))))).)).))))... ( -26.50) >DroSim_CAF1 3457 91 + 1 GUUCUCAGCAAUCAGGAGAACGUUUCGCCGCUUUCCGAUGAGAACGUUUCUUAG--CGCCUAUCGAUAACCAGUUAUCGAUGAUGGUGAGUCA (((((((.......(((((.((......)).)))))..))))))).........--(((((((((((((....)))))))))..))))..... ( -28.60) >DroEre_CAF1 3796 76 + 1 GUUCUCAGCAAUCAGCAGAACG---------------AUGAGAACGCUUCUCAG--CGCCUAUCGAUAACCAGUUAUCGAUGUUGGUGAUUCA (((((..((.....)))))))(---------------((((((.....))))).--(((((((((((((....)))))))))..))))..)). ( -23.40) >consensus GUUCUCAGCAAUCAGGAGAACGUUUCGCCGCUUUCCGAUGAGAACGUUUCUUAG__CGCCUAUCGAUAACCAGUUAUCGAUGAUGGUGAGUCA (((((((...............................)))))))...........(((((((((((((....)))))))))..))))..... (-19.70 = -19.96 + 0.25)

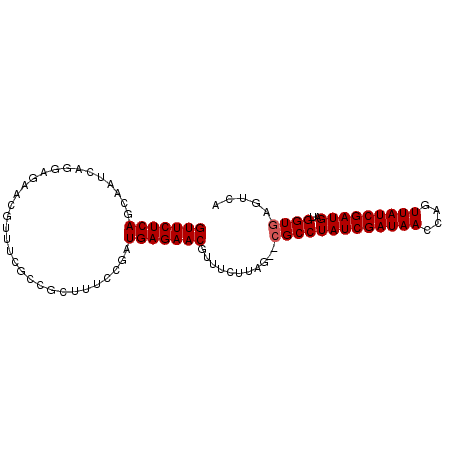
| Location | 3,015,726 – 3,015,827 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.92 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -20.12 |
| Energy contribution | -20.92 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3015726 101 + 22224390 G-UUUCGACGCUUUUCGAUGAGAACGUUUC--UUCGUUCGCCUAUCGAUAACCAGUUAUCGAUGAUGGUGAGUCAUGACUCAUGACUGGCCACGAGCCAAGUGG--- .-......(((((((((....(((((....--..)))))...(((((((((....))))))))).((((.(((((((...))))))).))))))))..))))).--- ( -36.60) >DroSec_CAF1 3539 99 + 1 G-UUUCGCUGCUUUCCGAUGAGAACGUUUC--UUAG--CGCCUAUCGAUAACCAGUUAUCGAUGAUGGUAAGUCAUGACUCAUGACUGGCCACGAGCCAAGUGG--- .-...((((((((..((.(((((.....))--))).--))..(((((((((....))))))))).((((.(((((((...))))))).)))).))))..)))).--- ( -31.00) >DroSim_CAF1 3478 99 + 1 G-UUUCGCCGCUUUCCGAUGAGAACGUUUC--UUAG--CGCCUAUCGAUAACCAGUUAUCGAUGAUGGUGAGUCAUGACUCAUGACUGGCCACGAGCCAAGUGG--- .-.....((((((.....(((((.....))--)))(--(...(((((((((....))))))))).((((.(((((((...))))))).))))...)).))))))--- ( -33.30) >DroEre_CAF1 3817 84 + 1 G----------------AUGAGAACGCUUC--UCAG--CGCCUAUCGAUAACCAGUUAUCGAUGUUGGUGAUUCAUGACUCAUGACUGGCCACGAGCCAAGAGG--- .----------------.(((((.....))--))).--(((((((((((((....)))))))))..))))..(((((...))))).((((.....)))).....--- ( -24.80) >DroAna_CAF1 3511 99 + 1 GCUUUUCACGCUUUCCGAUGAGAACGCUUCUCUCAG--CACCUAUCGAUAACCAGUUAUCGAUGCCGG------GUGAGUCAUGACUGGCCACGAGCCAAAAAAAAA .........((((.(((.(((((.......))))).--....(((((((((....))))))))).)))------(((.((((....))))))))))).......... ( -26.20) >consensus G_UUUCGACGCUUUCCGAUGAGAACGUUUC__UUAG__CGCCUAUCGAUAACCAGUUAUCGAUGAUGGUGAGUCAUGACUCAUGACUGGCCACGAGCCAAGUGG___ .........................((((.............(((((((((....)))))))))..(((.(((((((...))))))).)))..)))).......... (-20.12 = -20.92 + 0.80)

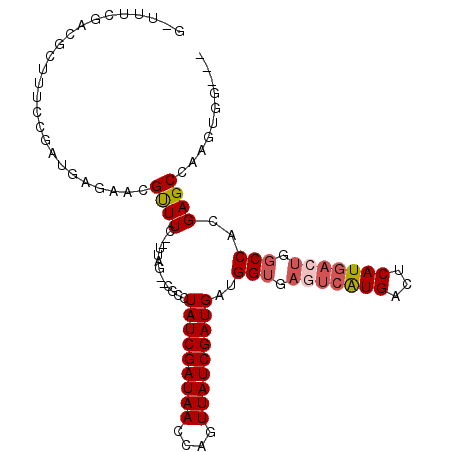
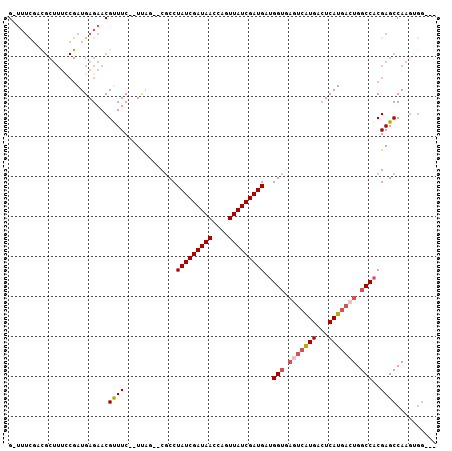
| Location | 3,015,726 – 3,015,827 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.92 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3015726 101 - 22224390 ---CCACUUGGCUCGUGGCCAGUCAUGAGUCAUGACUCACCAUCAUCGAUAACUGGUUAUCGAUAGGCGAACGAA--GAAACGUUCUCAUCGAAAAGCGUCGAAA-C ---.....((((((((((....))))))))))((((.(.((...((((((((....)))))))).)).(((((..--....)))))..........).))))...-. ( -30.50) >DroSec_CAF1 3539 99 - 1 ---CCACUUGGCUCGUGGCCAGUCAUGAGUCAUGACUUACCAUCAUCGAUAACUGGUUAUCGAUAGGCG--CUAA--GAAACGUUCUCAUCGGAAAGCAGCGAAA-C ---...((((((((((((..(((((((...)))))))..)))).((((((((....)))))))).)).)--))))--)...((((((........)).))))...-. ( -29.70) >DroSim_CAF1 3478 99 - 1 ---CCACUUGGCUCGUGGCCAGUCAUGAGUCAUGACUCACCAUCAUCGAUAACUGGUUAUCGAUAGGCG--CUAA--GAAACGUUCUCAUCGGAAAGCGGCGAAA-C ---...((((((((((((..(((((((...)))))))..)))).((((((((....)))))))).)).)--))))--)...((((.((....)).))))......-. ( -31.10) >DroEre_CAF1 3817 84 - 1 ---CCUCUUGGCUCGUGGCCAGUCAUGAGUCAUGAAUCACCAACAUCGAUAACUGGUUAUCGAUAGGCG--CUGA--GAAGCGUUCUCAU----------------C ---..((.((((((((((....)))))))))).)).........((((((((....)))))))).((((--((..--..)))))).....----------------. ( -29.20) >DroAna_CAF1 3511 99 - 1 UUUUUUUUUGGCUCGUGGCCAGUCAUGACUCAC------CCGGCAUCGAUAACUGGUUAUCGAUAGGUG--CUGAGAGAAGCGUUCUCAUCGGAAAGCGUGAAAAGC .......(((((.....)))))(((((.(((((------(....((((((((....)))))))).))))--((((((((.....)))).))))..)))))))..... ( -29.40) >consensus ___CCACUUGGCUCGUGGCCAGUCAUGAGUCAUGACUCACCAUCAUCGAUAACUGGUUAUCGAUAGGCG__CUAA__GAAACGUUCUCAUCGGAAAGCGGCGAAA_C ........((((((((((....)))))))))).......((...((((((((....)))))))).))........................................ (-20.52 = -20.72 + 0.20)

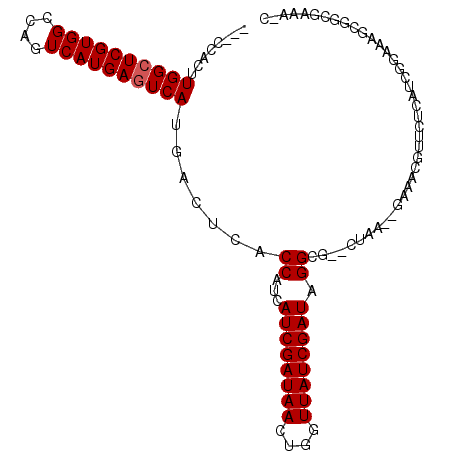
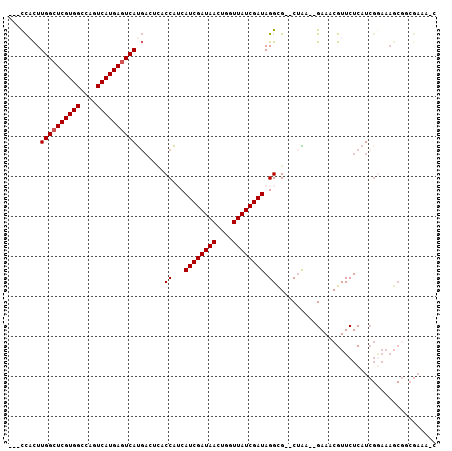
| Location | 3,015,763 – 3,015,859 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -20.22 |
| Energy contribution | -21.42 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
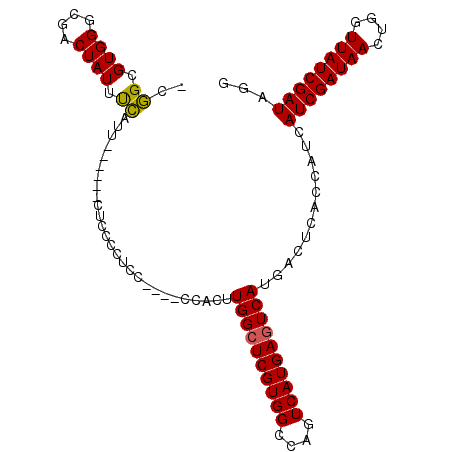
>X_DroMel_CAF1 3015763 96 + 22224390 CCUAUCGAUAACCAGUUAUCGAUGAUGGUGAGUCAUGACUCAUGACUGGCCACGAGCCAAGUGG---GGGAGGAGAG------ACCGAAAUAGUCGCCCACGCCG- ..(((((((((....))))))))).((((.(((((((...))))))).))))((.((...((((---(.((((....------.)).......)).)))))))))- ( -35.11) >DroSec_CAF1 3574 95 + 1 CCUAUCGAUAACCAGUUAUCGAUGAUGGUAAGUCAUGACUCAUGACUGGCCACGAGCCAAGUGG----GGAGGGGAG------AAUGAAAUAGUCGCCCACGCCG- (((((((((((....))))))))).((((.(((((((...))))))).))))..........))----((..(((.(------(.........)).)))...)).- ( -30.80) >DroSim_CAF1 3513 95 + 1 CCUAUCGAUAACCAGUUAUCGAUGAUGGUGAGUCAUGACUCAUGACUGGCCACGAGCCAAGUGG----GGAGGGGAG------AAUGGAAUAGUCGCCCACGCCG- (((((((((((....))))))))).((((.(((((((...))))))).))))..........))----((..(((.(------(.........)).)))...)).- ( -32.90) >DroEre_CAF1 3837 87 + 1 CCUAUCGAUAACCAGUUAUCGAUGUUGGUGAUUCAUGACUCAUGACUGGCCACGAGCCAAGAGG----G--------------AAAGAAAUAGUCGCCCACGCCG- ..(((((((((....)))))))))..((((..(((((...))))).((((.....))))...((----(--------------...((.....)).))).)))).- ( -26.10) >DroYak_CAF1 3630 101 + 1 CCUAUCGAUAACCAGUUAUCGAUGAUGGUGAGUCAUGACUCAUGACUGGCCACGAGCCAAGUGG----GGAGGGGACGGCGGUAAAGAAAUAGUCGCCCACGCCG- (((((((((((....))))))))).((((.(((((((...))))))).))))..........))----((..(((.((((............)))))))...)).- ( -38.00) >DroAna_CAF1 3549 94 + 1 CCUAUCGAUAACCAGUUAUCGAUGCCGG------GUGAGUCAUGACUGGCCACGAGCCAAAAAAAAAUAAAGGAAAA------AAUUAAAUAGUCGCCCACACAGU ..(((((((((....)))))))))..((------((((((....))((((.....))))..................------..........))))))....... ( -23.20) >consensus CCUAUCGAUAACCAGUUAUCGAUGAUGGUGAGUCAUGACUCAUGACUGGCCACGAGCCAAGUGG____GGAGGGGAG______AAUGAAAUAGUCGCCCACGCCG_ ..(((((((((....)))))))))..(((((((((((...))))))((((.....))))..................................)))))........ (-20.22 = -21.42 + 1.20)

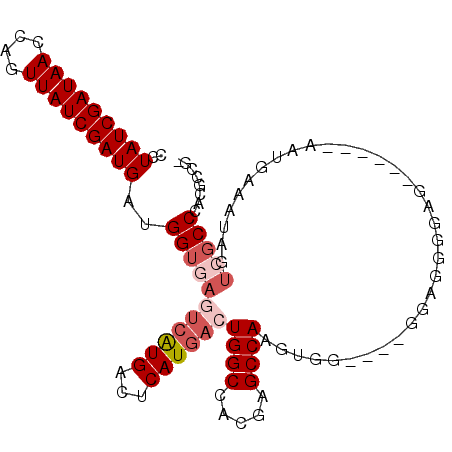
| Location | 3,015,763 – 3,015,859 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -28.59 |
| Consensus MFE | -25.15 |
| Energy contribution | -24.90 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3015763 96 - 22224390 -CGGCGUGGGCGACUAUUUCGGU------CUCUCCUCCC---CCACUUGGCUCGUGGCCAGUCAUGAGUCAUGACUCACCAUCAUCGAUAACUGGUUAUCGAUAGG -.((.(((((.((.......((.------....)))).)---)))).((((((((((....)))))))))).......))...((((((((....))))))))... ( -31.21) >DroSec_CAF1 3574 95 - 1 -CGGCGUGGGCGACUAUUUCAUU------CUCCCCUCC----CCACUUGGCUCGUGGCCAGUCAUGAGUCAUGACUUACCAUCAUCGAUAACUGGUUAUCGAUAGG -.((.(((((.((..........------......)))----)))).((((((((((....)))))))))).......))...((((((((....))))))))... ( -30.29) >DroSim_CAF1 3513 95 - 1 -CGGCGUGGGCGACUAUUCCAUU------CUCCCCUCC----CCACUUGGCUCGUGGCCAGUCAUGAGUCAUGACUCACCAUCAUCGAUAACUGGUUAUCGAUAGG -.((.(((((.((..........------......)))----)))).((((((((((....)))))))))).......))...((((((((....))))))))... ( -30.29) >DroEre_CAF1 3837 87 - 1 -CGGCGUGGGCGACUAUUUCUUU--------------C----CCUCUUGGCUCGUGGCCAGUCAUGAGUCAUGAAUCACCAACAUCGAUAACUGGUUAUCGAUAGG -.((.((((....)))).))...--------------.----((((.((((((((((....)))))))))).)).........((((((((....)))))))).)) ( -27.10) >DroYak_CAF1 3630 101 - 1 -CGGCGUGGGCGACUAUUUCUUUACCGCCGUCCCCUCC----CCACUUGGCUCGUGGCCAGUCAUGAGUCAUGACUCACCAUCAUCGAUAACUGGUUAUCGAUAGG -.((.(.(((.(((...............)))))).).----)).(.((((((((((....)))))))))).)..........((((((((....))))))))... ( -31.66) >DroAna_CAF1 3549 94 - 1 ACUGUGUGGGCGACUAUUUAAUU------UUUUCCUUUAUUUUUUUUUGGCUCGUGGCCAGUCAUGACUCAC------CCGGCAUCGAUAACUGGUUAUCGAUAGG .(((.((((((((((...(((..------.......))).........(((.....)))))))..).)))))------.))).((((((((....))))))))... ( -21.00) >consensus _CGGCGUGGGCGACUAUUUCAUU______CUCCCCUCC____CCACUUGGCUCGUGGCCAGUCAUGAGUCAUGACUCACCAUCAUCGAUAACUGGUUAUCGAUAGG ..((.((((....)))).))...........................((((((((((....))))))))))............((((((((....))))))))... (-25.15 = -24.90 + -0.25)
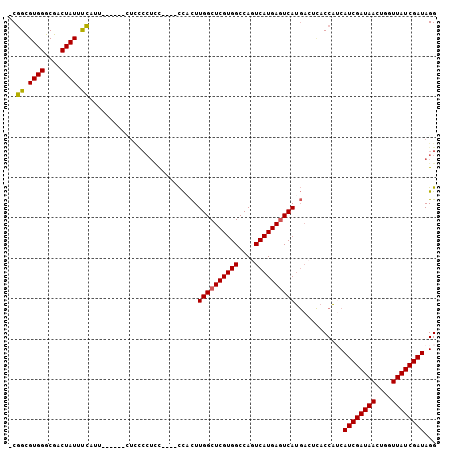
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:46 2006