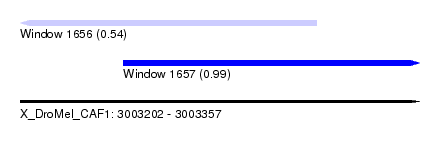
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,003,202 – 3,003,357 |
| Length | 155 |
| Max. P | 0.988774 |

| Location | 3,003,202 – 3,003,317 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -22.87 |
| Energy contribution | -23.53 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
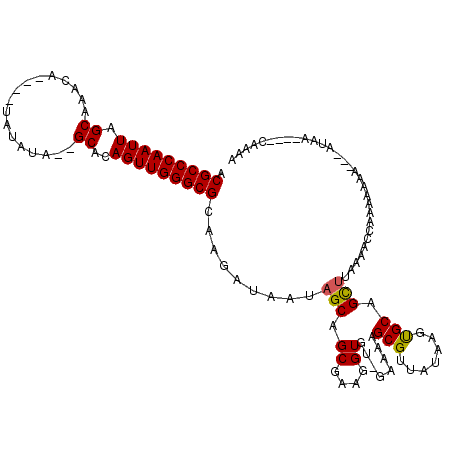
>X_DroMel_CAF1 3003202 115 - 22224390 AUAACGCUUUUCAACACCUUCGCUGCUAAUAUCUUGCGCCCAACUGUGCUCUAUAUA----UGUUUGCUAAUUGGGCGUGUGGCGAUUUCCUGCAAAGCUGCAAAGAGGUCUGAAUACA .........((((..((((((((..(.........((((((((.((.((........----.....)))).)))))))))..)))).....((((....))))...)))).)))).... ( -29.62) >DroEre_CAF1 411916 97 - 1 AUAACGCUUUUC-ACACCUUCGCUGCUAUUAUUUUGCGCCCAACUGUGC--UAUAUA----UGUUUGCUAAUUGGGCGUGUGGCCAUUUCCUGC---GCUGGAAAAA------------ ..........((-((((.......((.........))((((((.((.((--......----.....)))).))))))))))))...(((((...---...)))))..------------ ( -23.60) >DroYak_CAF1 393157 101 - 1 AUAACGCUUUUC-ACACCUUCGCUGCUAUUCUCUUGCGCCCAACUGUGC--UAUAUAUAUGUGUUUGCUAAUUGGGCGUGUGGCGAUUUCCUGC---GCUGGAAAAA------------ ............-......((((..(.........((((((((.((.((--.((((....))))..)))).)))))))))..))))(((((...---...)))))..------------ ( -27.90) >consensus AUAACGCUUUUC_ACACCUUCGCUGCUAUUAUCUUGCGCCCAACUGUGC__UAUAUA____UGUUUGCUAAUUGGGCGUGUGGCGAUUUCCUGC___GCUGGAAAAA____________ ...................((((..(.........((((((((.((.((...(((......)))..)))).)))))))))..))))(((((.((...)).))))).............. (-22.87 = -23.53 + 0.67)


| Location | 3,003,242 – 3,003,357 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -17.93 |
| Energy contribution | -17.80 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3003242 115 + 22224390 ACGCCCAAUUAGCAAACA----UAUAUAGAGCACAGUUGGGCGCAAGAUAUUAGCAGCGAAGGUGUUGAAAAGCGUUAUGAGUGCAGCUUUAAACCAAAAAAGAAAAAAAAAAUCGAAA .(((((((((.((.....----........))..)))))))))...(((.((((((.(....)))))))(((((((.......)).))))).....................))).... ( -23.52) >DroSec_CAF1 380196 102 + 1 ACGCCCAAUUAGCAAACA----UAUAUA--GCACAGUUGGGCGCAAGAUAUUAGCAGCGAAGGUGU-GAAGAGCGUUAUGAGUGCAGUG------CAAUAAUGCAAAUAA----CAAAA .(((((((((.((.....----......--))..)))))))))...(......(((.(....))))-......)(((((...((((...------......)))).))))----).... ( -23.70) >DroEre_CAF1 411941 101 + 1 ACGCCCAAUUAGCAAACA----UAUAUA--GCACAGUUGGGCGCAAAAUAAUAGCAGCGAAGGUGU-GAAAAGCGUUAUAAGCGCAGCUUAAAGCUAUAAAAA-----------UAAAG .(((((((((.((.....----......--))..))))))))).......(((((.((....))..-.....((((.....))))........))))).....-----------..... ( -26.00) >DroYak_CAF1 393182 109 + 1 ACGCCCAAUUAGCAAACACAUAUAUAUA--GCACAGUUGGGCGCAAGAGAAUAGCAGCGAAGGUGU-GAAAAGCGUUAUAAGUGCCGCUAAAAACCAUUAAAA---AUGC----CAAAA .(((((((((.((...............--))..)))))))))........((((.(((....(((-((......)))))..))).)))).............---....----..... ( -22.46) >consensus ACGCCCAAUUAGCAAACA____UAUAUA__GCACAGUUGGGCGCAAGAUAAUAGCAGCGAAGGUGU_GAAAAGCGUUAUAAGUGCAGCUUAAAACCAAAAAAA___AUAA____CAAAA .(((((((((.((.................))..))))))))).........(((.((....))........(((.......))).))).............................. (-17.93 = -17.80 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:37 2006