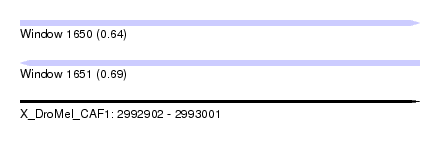
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,992,902 – 2,993,001 |
| Length | 99 |
| Max. P | 0.693819 |

| Location | 2,992,902 – 2,993,001 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
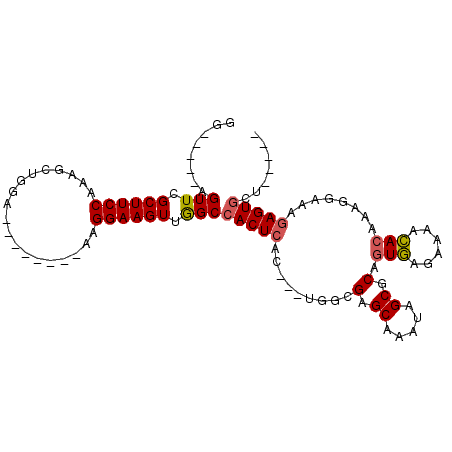
| Reading direction | forward |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -19.38 |
| Consensus MFE | -14.30 |
| Energy contribution | -14.68 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2992902 99 + 22224390 CUCUAGGCACUCUUUCCUUUGUGUUUUCUCACUGCGCUAUUUGCUCGCUAGUAGUGAGUGGCCAACUUCCUU--------UCCAGCUUUGGAAGCGAACUCUCCUCC .....(((((((........(((......)))...((((((.(....).)))))))))).)))...(((.((--------(((......))))).)))......... ( -21.80) >DroSec_CAF1 370085 86 + 1 -----AGCACUCUUUCCUUUGUGUUUUCUCACUGCGCUAUUUGCUCGCCA---GUGAGUGGCUAACUUCCUU--------UCCAGCUUUGGAAGCGAACU-----CC -----(((((..........)))))..(((((((.((.........))))---)))))........(((.((--------(((......))))).)))..-----.. ( -20.00) >DroSim_CAF1 335300 86 + 1 -----AGCACUCUUUCCUUUGUGUUUUCUCACUGCGCUAUUUGCUCGCCA---GUGAGUGGCCAACUUCCUU--------UCCAGCUUUGGAAGCGAACU-----CC -----.((..........(((.(((..(((((((.((.........))))---))))).))))))......(--------(((......)))))).....-----.. ( -20.70) >DroYak_CAF1 382517 89 + 1 CUU----UACUUUUU-UUUUUUAUUUUUUUACUGCGCUAUUUGCUCUG-------GAGUGGCCAACUUCCU-UCCAACUUUCCAGCUUUGGAAGCGAACU-----CC ...----........-................((.(((((((......-------)))))))))..(((((-(((((..........))))))).)))..-----.. ( -15.00) >consensus _____AGCACUCUUUCCUUUGUGUUUUCUCACUGCGCUAUUUGCUCGCCA___GUGAGUGGCCAACUUCCUU________UCCAGCUUUGGAAGCGAACU_____CC ....................(((......)))..((((..(..(((((.....)))))..)...................(((......)))))))........... (-14.30 = -14.68 + 0.37)

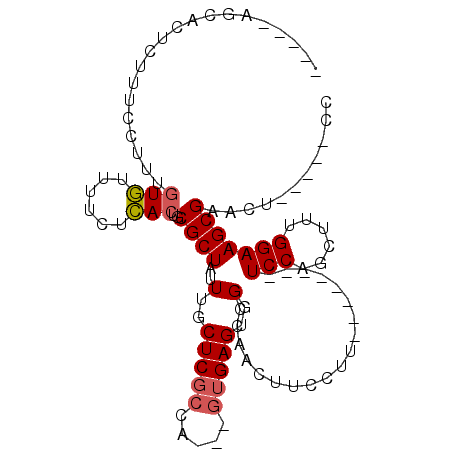
| Location | 2,992,902 – 2,993,001 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2992902 99 - 22224390 GGAGGAGAGUUCGCUUCCAAAGCUGGA--------AAGGAAGUUGGCCACUCACUACUAGCGAGCAAAUAGCGCAGUGAGAAAACACAAAGGAAAGAGUGCCUAGAG (((((.(....).)))))..((((...--------.....))))(((.((((.......(((.........))).(((......)))........)))))))..... ( -22.90) >DroSec_CAF1 370085 86 - 1 GG-----AGUUCGCUUCCAAAGCUGGA--------AAGGAAGUUAGCCACUCAC---UGGCGAGCAAAUAGCGCAGUGAGAAAACACAAAGGAAAGAGUGCU----- ((-----..(((..((((...(((((.--------.......)))))..(((((---((((.........)).)))))))..........)))).)))..))----- ( -23.00) >DroSim_CAF1 335300 86 - 1 GG-----AGUUCGCUUCCAAAGCUGGA--------AAGGAAGUUGGCCACUCAC---UGGCGAGCAAAUAGCGCAGUGAGAAAACACAAAGGAAAGAGUGCU----- ((-----..(((..((((...((..(.--------.......)..))..(((((---((((.........)).)))))))..........)))).)))..))----- ( -22.90) >DroYak_CAF1 382517 89 - 1 GG-----AGUUCGCUUCCAAAGCUGGAAAGUUGGA-AGGAAGUUGGCCACUC-------CAGAGCAAAUAGCGCAGUAAAAAAAUAAAAAA-AAAAAGUA----AAG ((-----(((((((((((..((((....))))...-.))))).)))..))))-------).(.((.....)).).................-........----... ( -19.40) >consensus GG_____AGUUCGCUUCCAAAGCUGGA________AAGGAAGUUGGCCACUCAC___UGGCGAGCAAAUAGCGCAGUGAGAAAACACAAAGGAAAGAGUGCU_____ ........(((.((((((...................)))))).)))(((((.........(.((.....)).).(((......)))........)))))....... (-14.15 = -14.33 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:32 2006