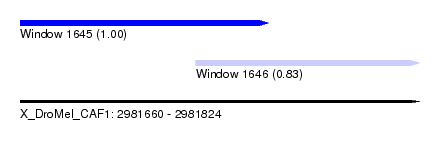
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,981,660 – 2,981,824 |
| Length | 164 |
| Max. P | 0.998926 |

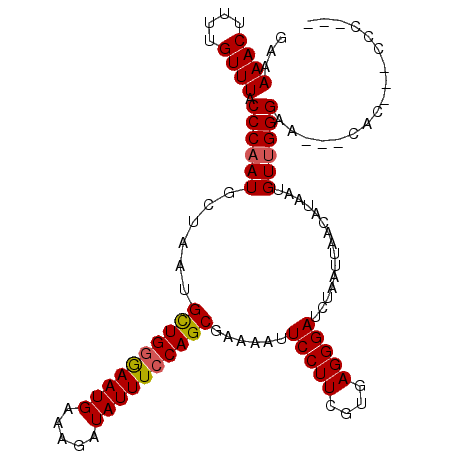
| Location | 2,981,660 – 2,981,762 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.73 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.43 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2981660 102 + 22224390 GAAAAACUUUUGUUUACCCAAUGCUAAUGCUGGGAAUGAAAGAUAUUUCCAGCGAAAAUUCCUUCGUGAGGGAUCUAAUUAACAUAAUGUUGGGAA---CAC---CCC--- ...((((....)))).((((((..((.(((((((((((.....))))))))))).....(((((....)))))...........))..))))))..---...---...--- ( -26.90) >DroSec_CAF1 359272 102 + 1 GAAAAACUUUUGUUUACCCAAUGCUAAUGCUGGGAAUGAAAGAUAUUUCCAGCGAAAAUUCCUUCGUGAGGGAUCUAAUUAACAUAAUGUUGGGAA---CAC---CCC--- ...((((....)))).((((((..((.(((((((((((.....))))))))))).....(((((....)))))...........))..))))))..---...---...--- ( -26.90) >DroSim_CAF1 325005 102 + 1 GAAAAACUUUUGUUUACCCAAUGCUAAUGCUGGAAAUGAAAGAUAUUUCCAGCGAAAAUUCCUUCGUGAGGGAUCUAAUUAACAUAAUGUUGGGAA---CAC---CCC--- ...((((....)))).((((((..((.(((((((((((.....))))))))))).....(((((....)))))...........))..))))))..---...---...--- ( -27.40) >DroEre_CAF1 390167 108 + 1 GAAAAACUUUUGUUUACCCCAUGCUAAUGCUGGGAAUGAAAGAUAUUUCCAGCGAAAAUUCCUUCGUGAGGGAUCUAAUUAACAUAAUGUUGGGAA---CACACCCUUCCC ...((((....)))).((((((......((((((((((.....))))))))))(((......)))))).)))...................(((((---.......))))) ( -25.20) >DroYak_CAF1 371012 105 + 1 GAAAAACUUUUGUUUACCCAAUGCUAAUGCUGGGAAUGAAAGAUAUUUCCAGCGAAAAUUCCUUCGUGAGGGAUCUAAUUAACAUAAUGUUGGGAA---CAC---CACCCC ...((((....)))).((((((..((.(((((((((((.....))))))))))).....(((((....)))))...........))..))))))..---...---...... ( -26.90) >DroAna_CAF1 557885 101 + 1 GAAAAACUUUUGUUUACCCAAU------GUUGAGAAUGAAAGAUAUUUCCAGCGAAAA-UCCUUCGUGAGGGAUCUAAUUAACAUAAUGUUGGGAAAGACAC---CCACAC ..........(((((.((((((------((((..(((...(((...((((.(((((..-...)))))..))))))).)))....))))))))))..))))).---...... ( -20.60) >consensus GAAAAACUUUUGUUUACCCAAUGCUAAUGCUGGGAAUGAAAGAUAUUUCCAGCGAAAAUUCCUUCGUGAGGGAUCUAAUUAACAUAAUGUUGGGAA___CAC___CCC___ ...((((....)))).((((((......((((((((((.....))))))))))......(((((....)))))...............))))))................. (-24.38 = -24.43 + 0.06)

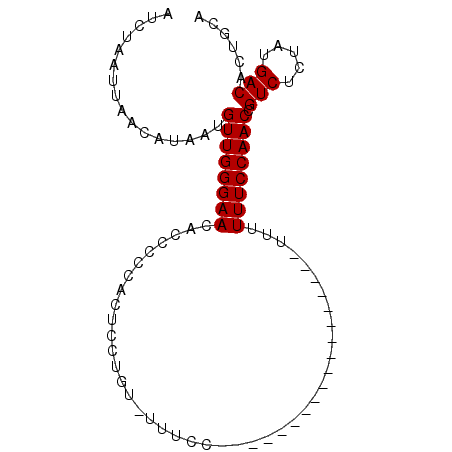
| Location | 2,981,732 – 2,981,824 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -9.77 |
| Consensus MFE | -8.15 |
| Energy contribution | -8.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2981732 92 + 22224390 AUCUAAUUAACAUAAUGUUGGGAACACCCCCACUCCUGGUUUUCCUUUUCCUUUUCCUUCUUUUUUUUUCCAACCGUCUCUAUGACACUGCA .......((((.....))))((((.(((.........))).))))..............................(((.....)))...... ( -9.80) >DroSec_CAF1 359344 73 + 1 AUCUAAUUAACAUAAUGUUGGGAACACCCCCACUUCUGU-UUUCC------------------UUUUUUCCAACCGUCUCUAUGACACUGCA ................((((((((.......((....))-.....------------------...)))))))).(((.....)))...... ( -9.76) >DroSim_CAF1 325077 73 + 1 AUCUAAUUAACAUAAUGUUGGGAACACCCCCACUCCUGU-UUUCC------------------UUUUUUCCAACCGUCUCUAUGACACUGCA ................((((((((.......((....))-.....------------------...)))))))).(((.....)))...... ( -9.76) >consensus AUCUAAUUAACAUAAUGUUGGGAACACCCCCACUCCUGU_UUUCC__________________UUUUUUCCAACCGUCUCUAUGACACUGCA ................((((((((..........................................)))))))).(((.....)))...... ( -8.15 = -8.15 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:28 2006