| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,973,257 – 2,973,351 |
| Length | 94 |
| Max. P | 0.679428 |

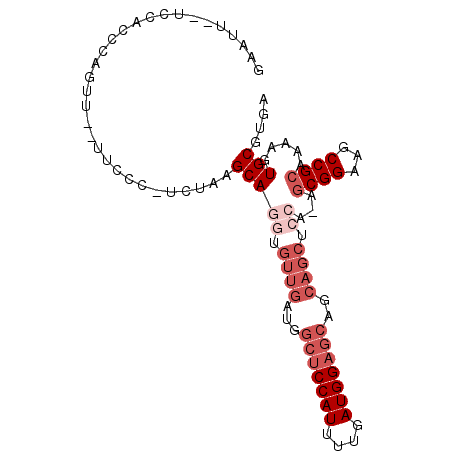
| Location | 2,973,257 – 2,973,351 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -17.48 |
| Energy contribution | -18.77 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2973257 94 + 22224390 UCACGCACUUUUGCGGCUUCCGCUUUGGAGCUGCUGCUCCAUCAAAAUGGAGCCAUCAACACCUGCUUAGA-GGUAAAAAAGUGGGUGGC--AGUUC ((((.(((((((((((((((......)))))))).(((((((....))))))).......((((......)-)))..))))))).)))).--..... ( -37.10) >DroGri_CAF1 509383 80 + 1 UCACGCACUUUUGCGGCUUCCGGC-UGG----------CCAUCAAAAUGGAGCCAUCAACACCUGCUUAGG-GAUCG--G-CAGGCGCCA--GGCAU ...((((....))))((((..(((-(((----------(..((......))))))......((((((....-....)--)-)))).))))--))).. ( -24.90) >DroSec_CAF1 350772 91 + 1 UCACGCACUUUUGCGGCUUCCGCU-UGGAGCUGCUGCUCCAUCAAAAUGGAGCCAUCAACACCUGCUUAGA-GGGAA--AACUGGGUGGA--AAUCC ((((.((.((((((((((((....-.)))))))).(((((((....)))))))........(((......)-)).))--)).)).)))).--..... ( -30.40) >DroSim_CAF1 317001 91 + 1 UCACGCACUUUUGCGGCUUCCGCU-UGGAGCUGCUGCUCCAUCAAAAUGGAGCCAUCAACACCUGCUUAGA-GGGAA--AACUGGGUGGA--AAUUC ((((.((.((((((((((((....-.)))))))).(((((((....)))))))........(((......)-)).))--)).)).)))).--..... ( -30.40) >DroEre_CAF1 380204 83 + 1 UCACGCACUUUUGCGGCUUCCGCU-U--AGCUGCUGCUCCAUCAAAAUGGAGCCAUCAACACCUGCUUAGA-GGGAA--AACUCUGUGU-------- .((((((.....((((((......-.--)))))).(((((((....)))))))..........)))..(((-(....--..))))))).-------- ( -26.90) >DroYak_CAF1 361436 92 + 1 UCACGCACUUUUGCGGCUUCCGCU-U--AGCUGCUGCUCCAUCAAAAUGGAGCCAUCAACACCUGCUUAGAAGGGAA--AACUGAGUGGGAAAACUC ...((((....))))..(((((((-(--((.....(((((((....)))))))........(((.......)))...--..))))))))))...... ( -30.00) >consensus UCACGCACUUUUGCGGCUUCCGCU_UGGAGCUGCUGCUCCAUCAAAAUGGAGCCAUCAACACCUGCUUAGA_GGGAA__AACUGGGUGGA__AAUUC .((((((....)))((((((.....((((((....)))))).......))))))...............................)))......... (-17.48 = -18.77 + 1.29)

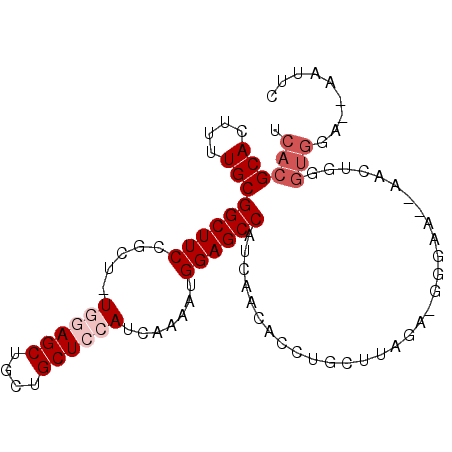
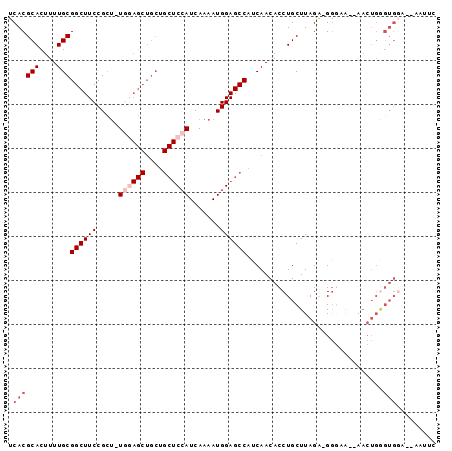
| Location | 2,973,257 – 2,973,351 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -20.08 |
| Energy contribution | -22.08 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2973257 94 - 22224390 GAACU--GCCACCCACUUUUUUACC-UCUAAGCAGGUGUUGAUGGCUCCAUUUUGAUGGAGCAGCAGCUCCAAAGCGGAAGCCGCAAAAGUGCGUGA .....--..(((.(((((((.....-........((.((((...(((((((....)))))))..)))).))...((((...))))))))))).))). ( -34.80) >DroGri_CAF1 509383 80 - 1 AUGCC--UGGCGCCUG-C--CGAUC-CCUAAGCAGGUGUUGAUGGCUCCAUUUUGAUGG----------CCA-GCCGGAAGCCGCAAAAGUGCGUGA (((((--(.(((((((-(--.....-.....))))))(((..(((((((((....))))----------..)-))))..))).))...)).)))).. ( -27.70) >DroSec_CAF1 350772 91 - 1 GGAUU--UCCACCCAGUU--UUCCC-UCUAAGCAGGUGUUGAUGGCUCCAUUUUGAUGGAGCAGCAGCUCCA-AGCGGAAGCCGCAAAAGUGCGUGA ((.((--(((.....(((--(....-...)))).((.((((...(((((((....)))))))..)))).)).-...)))))))(((....))).... ( -31.20) >DroSim_CAF1 317001 91 - 1 GAAUU--UCCACCCAGUU--UUCCC-UCUAAGCAGGUGUUGAUGGCUCCAUUUUGAUGGAGCAGCAGCUCCA-AGCGGAAGCCGCAAAAGUGCGUGA .....--..(((.((.((--((...-........((.((((...(((((((....)))))))..)))).)).-.((((...)))))))).)).))). ( -30.90) >DroEre_CAF1 380204 83 - 1 --------ACACAGAGUU--UUCCC-UCUAAGCAGGUGUUGAUGGCUCCAUUUUGAUGGAGCAGCAGCU--A-AGCGGAAGCCGCAAAAGUGCGUGA --------.(((((((..--....)-)))..(((...((((...(((((((....)))))))..)))).--.-.((((...)))).....)))))). ( -30.20) >DroYak_CAF1 361436 92 - 1 GAGUUUUCCCACUCAGUU--UUCCCUUCUAAGCAGGUGUUGAUGGCUCCAUUUUGAUGGAGCAGCAGCU--A-AGCGGAAGCCGCAAAAGUGCGUGA ((((......))))....--...........(((...((((...(((((((....)))))))..)))).--.-.((((...)))).....))).... ( -27.30) >consensus GAAUU__UCCACCCAGUU__UUCCC_UCUAAGCAGGUGUUGAUGGCUCCAUUUUGAUGGAGCAGCAGCUCCA_AGCGGAAGCCGCAAAAGUGCGUGA ...............................(((((.((((...(((((((....)))))))..)))).))...((((...)))).....))).... (-20.08 = -22.08 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:24 2006