| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,909,357 – 2,909,469 |
| Length | 112 |
| Max. P | 0.975448 |

| Location | 2,909,357 – 2,909,469 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.51 |
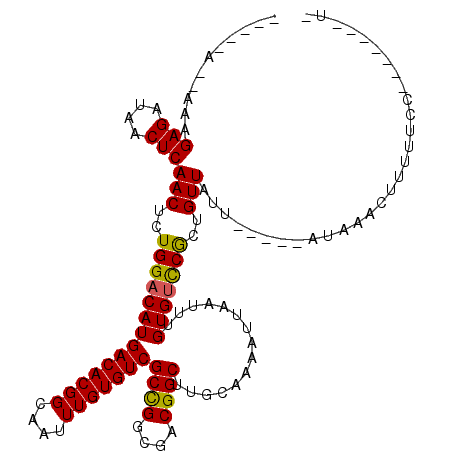
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -27.68 |
| Energy contribution | -27.27 |
| Covariance contribution | -0.42 |
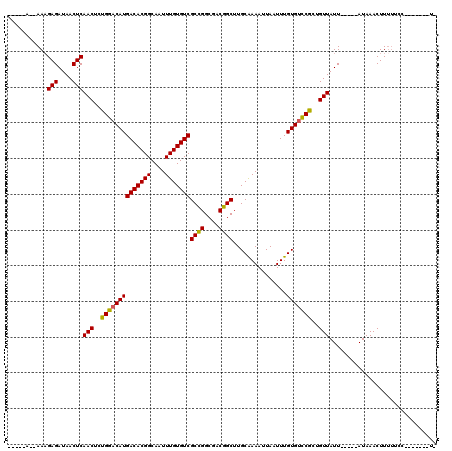
| Combinations/Pair | 1.12 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
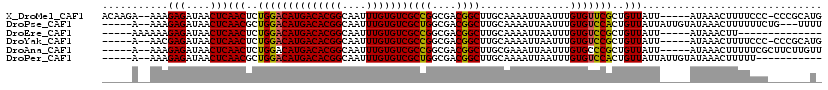
>X_DroMel_CAF1 2909357 112 - 22224390 ACAAGA--AAAGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCAAAAUUAAUUUGUGUUCGCUGUUAUU-----AUAAACUUUUCCC-CCCGCAUG ....((--((((..((((...(((..((((((((((((((....)))))))((((....))))...............)))))))..))).))-----))...))))))..-........ ( -29.40) >DroPse_CAF1 403762 110 - 1 -----A--AAAGAGAUAACUCAACGCUGGACAUGACACGGCAAUUUGUGUCGCUGGCGACGGCUUGCAAAAUUAAUUUGUGUCCACUGUUAUUAUUGUAUAAACUUUUUUCUG---UUUU -----.--...(((....)))((((.((((((((((((((....)))))))((((....))))...............))))))).))))..........((((........)---))). ( -28.50) >DroEre_CAF1 320026 96 - 1 -----AAAAAAGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCAAAAUUAAUUUGUGUCCGCUGUUAUU-----AUAAACUU-------------- -----......(((....)))(((..((((((((((((((....)))))))((((....))))...............)))))))..)))...-----........-------------- ( -29.20) >DroYak_CAF1 304580 107 - 1 -----A--AACGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCAAAAUUAAUUUGUGUCCGCUGUUAUU-----AUAAACUUUUCCC-CCCGCAUG -----.--...(((....)))(((..((((((((((((((....)))))))((((....))))...............)))))))..)))...-----.............-........ ( -29.30) >DroAna_CAF1 466691 108 - 1 -----A--AAAGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCGAAAUUAAUUUGUGCCCGCUGUUAUU-----AUAAACUUUUUCGCUUCUUGUU -----.--...(((....))).......((((.(((((((....)))))))((((....))))..((((((....((((((...........)-----)))))...))))))....)))) ( -25.30) >DroPer_CAF1 424238 102 - 1 -----A--AAAGAGAUAACUCAACGCUGGACAUGACACGGCAAUUUGUGUCGCUGGCGACGGCUUGCAAAAUUAAUUUGUGUCCACUGUUAUUAUUGUAUAAACUUUUU----------- -----.--...(((....)))((((.((((((((((((((....)))))))((((....))))...............))))))).))))...................----------- ( -28.30) >consensus _____A__AAAGAGAUAACUCAACUCUGGACAUGACACGGCAAUUUGUGUCGCCGGCGACGGCUUGCAAAAUUAAUUUGUGUCCGCUGUUAUU_____AUAAACUUUUUCC_______U_ ...........(((....)))(((..((((((((((((((....)))))))((((....))))...............)))))))..))).............................. (-27.68 = -27.27 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:13 2006