| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,908,663 – 2,908,760 |
| Length | 97 |
| Max. P | 0.933341 |

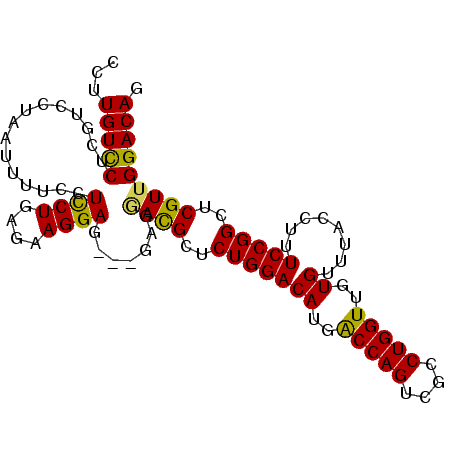
| Location | 2,908,663 – 2,908,760 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -28.17 |
| Energy contribution | -27.42 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
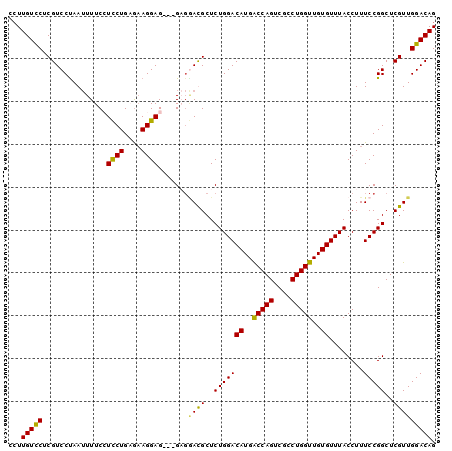
>X_DroMel_CAF1 2908663 97 + 22224390 CCUUGUCCUCGUCCUAAUUUUCGUCCUGCAAAGGAG---GCGGACGCUCUGGACAUGACCAGUCGCCUGGUUGUGUUUACCUUUCCGGCUCGUUGGACAG ...(((((.((............((((....))))(---.((((.....((((((..(((((....)))))..))))))....)))).).))..))))). ( -33.90) >DroSim_CAF1 264388 100 + 1 CCUUGUCCUCGUCCCAAUUUUCGUCCUGCGAAGGAGGAGGAGGACGCUCUGGACAUGGCCAGUCGCCUGGUUGUGUUUACCUUUCCGGCUCGUUGGACAG ...(((((.((..((..(((((.((((....)))).)))))(((.....((((((..(((((....)))))..))))))....)))))..))..))))). ( -40.50) >DroEre_CAF1 319340 97 + 1 CCUUGUUCACGUCCUAAUUUUCCUUCUGAGAAGGAG---GAGAAUGCUCUGGACAUGACCAGUCGCCUGGUUGUGUUUACCUUUCCGGCUCGUUGGACAG ...((((((((.((..(((((((((((....)))))---))))))....((((((..(((((....)))))..)))))).......))..)).)))))). ( -37.10) >DroYak_CAF1 303876 92 + 1 --UUGUCCUCGUCCUAAAUUUCCUCCUGAGCAGGAG---G---AUGCUCUGGACAUGGCCAGUCGCCUGGUUGUGUUUACCUUUCCGGCUCGUUGGACAG --.(((((.((.((......(((((((....)))))---)---).....((((((..(((((....)))))..)))))).......))..))..))))). ( -37.20) >consensus CCUUGUCCUCGUCCUAAUUUUCCUCCUGAGAAGGAG___GAGGACGCUCUGGACAUGACCAGUCGCCUGGUUGUGUUUACCUUUCCGGCUCGUUGGACAG ...(((((...............((((....)))).......((((..(((((((..(((((....)))))..))........)))))..))))))))). (-28.17 = -27.42 + -0.75)

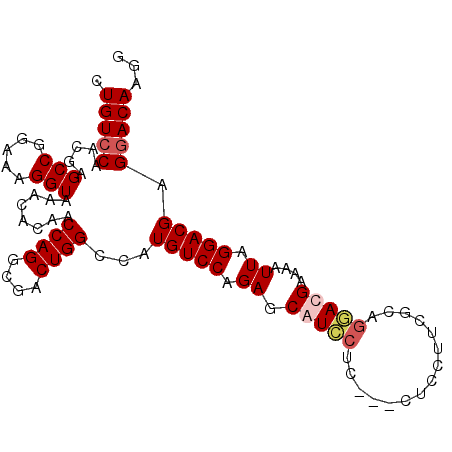
| Location | 2,908,663 – 2,908,760 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -22.93 |
| Energy contribution | -23.93 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2908663 97 - 22224390 CUGUCCAACGAGCCGGAAAGGUAAACACAACCAGGCGACUGGUCAUGUCCAGAGCGUCCGC---CUCCUUUGCAGGACGAAAAUUAGGACGAGGACAAGG .(((((.....(((.....))).......(((((....)))))..(((((.((.(((((((---.......)).)))))....)).))))).)))))... ( -33.60) >DroSim_CAF1 264388 100 - 1 CUGUCCAACGAGCCGGAAAGGUAAACACAACCAGGCGACUGGCCAUGUCCAGAGCGUCCUCCUCCUCCUUCGCAGGACGAAAAUUGGGACGAGGACAAGG (((..((....(((.....)))........((((....))))...))..)))...((((((.((((..((((.....))))....)))).)))))).... ( -31.90) >DroEre_CAF1 319340 97 - 1 CUGUCCAACGAGCCGGAAAGGUAAACACAACCAGGCGACUGGUCAUGUCCAGAGCAUUCUC---CUCCUUCUCAGAAGGAAAAUUAGGACGUGAACAAGG ((.(((........))).))..........((((....))))((((((((.(((....)))---.((((((...))))))......))))))))...... ( -29.40) >DroYak_CAF1 303876 92 - 1 CUGUCCAACGAGCCGGAAAGGUAAACACAACCAGGCGACUGGCCAUGUCCAGAGCAU---C---CUCCUGCUCAGGAGGAAAUUUAGGACGAGGACAA-- .(((((.....(((.....)))........((((....))))...(((((.(((..(---(---(((((....)))))))..))).))))).))))).-- ( -36.30) >consensus CUGUCCAACGAGCCGGAAAGGUAAACACAACCAGGCGACUGGCCAUGUCCAGAGCAUCCUC___CUCCUUCGCAGGACGAAAAUUAGGACGAGGACAAGG .(((((.....(((.....)))........((((....))))...(((((.((.(((((...............)))))....)).))))).)))))... (-22.93 = -23.93 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:12 2006