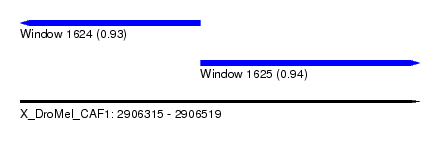
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,906,315 – 2,906,519 |
| Length | 204 |
| Max. P | 0.937213 |

| Location | 2,906,315 – 2,906,407 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -19.91 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2906315 92 - 22224390 CAGUGCCACUUUUUAGAGGUUUGCCAAGAACUUCUUCUAUCCGAAGUAGAA-AAACUUUUAGGGCACUGAAAAAUACGUUUU-UUAUAAUAAAU (((((((........(((((((.....)))))))((((((.....))))))-..........))))))).............-........... ( -22.60) >DroSec_CAF1 292974 92 - 1 CAGUGCCACUUUUUAGAGGUUUGCCAAGAACUUCUUCUACACGAAGCAGAA-AAACUUUUAGGGCACUGAAAAAUGCGUUUU-UUUUAAUAAGU (((((((.......((((((((.....)))))))).(((...((((.....-...)))))))))))))).............-........... ( -20.40) >DroSim_CAF1 262111 94 - 1 CAGUGCCACUUUUUAGAGGUUUGCCAAGAACUUCUUCUACACGAAGCAGAAAAAACUUUUAGGGCACUGAAAAAUGCGUUUUUUUUUAAUAAAU (((((((........(((((((.....)))))))((((.(.....).))))...........)))))))......................... ( -20.10) >DroEre_CAF1 316916 91 - 1 CAGUGCCACUUUUUAGAGCUUUGUCAAGAACUUGUUCUACCCAAAACAAGA-AAACUUUUCGGGCACUGAAAAAUAUGUUU--UAAUAAUAGAU (((((((......((((((...((.....))..))))))..........((-(.....))).)))))))............--........... ( -18.30) >DroYak_CAF1 301461 90 - 1 CAGUGCCACUUUUUAGAGGUUUGCCAAGAACUUCUUCUACCCAAAACAA-A-CAACUUUUAGGGCACUGAAAAAUACGUUU--UUAUAAUCAAU (((((((.((....((((((((.....))))))))..............-.-........)))))))))............--........... ( -18.13) >consensus CAGUGCCACUUUUUAGAGGUUUGCCAAGAACUUCUUCUACCCGAAGCAGAA_AAACUUUUAGGGCACUGAAAAAUACGUUUU_UUAUAAUAAAU (((((((.......((((((((.....))))))))....................(.....))))))))......................... (-15.68 = -16.08 + 0.40)

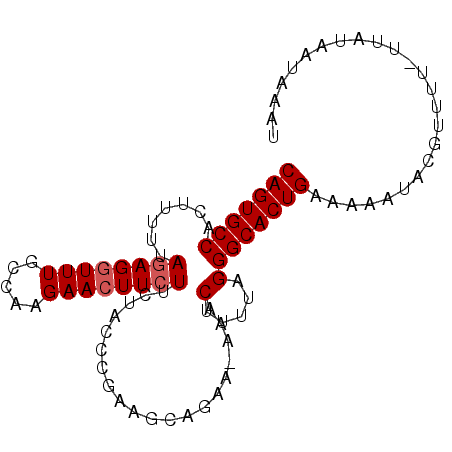
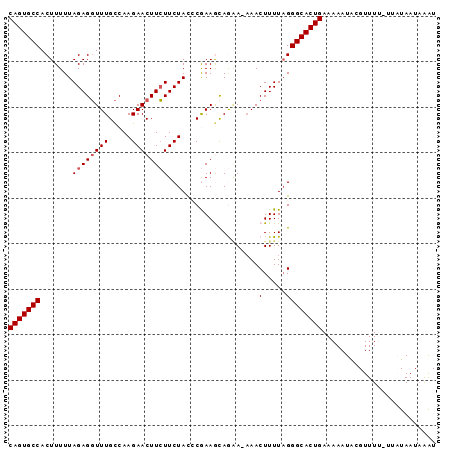
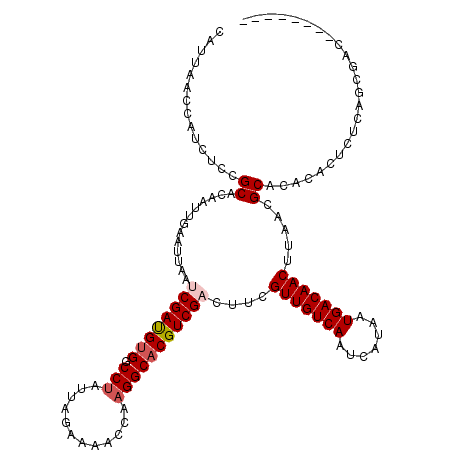
| Location | 2,906,407 – 2,906,519 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -21.41 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2906407 112 + 22224390 CAUUAACCAUCUCCGCACAAUUGAAUUAAUCGAUGUGGCCUAUUAGAAAACCAAGGCACGUCGACUUCGUUGUCAAUCAUAAUGACAACUUAACGCACACACUCUCAGCGAC-------- .............(((.............((((((((.(((............)))))))))))....(((((((.......)))))))..................)))..-------- ( -25.70) >DroSec_CAF1 293066 112 + 1 CAUUAACCAUCUCCGCACAAUUGAAUUAAUCGAUGUGGCCUAUUAGAAAACCAAGGCACGUCGUCUUCGUUGUCAAUCAUAAUGACAACUUAACGCACACACUCUCAGCGAC-------- .............(((......(((.....(((((((.(((............))))))))))..)))(((((((.......)))))))..................)))..-------- ( -24.30) >DroSim_CAF1 262205 112 + 1 CAUUAACCAUCUCCGCACAAUUGAAUUAAUCGAUGUGGCCUAUUAGAAAACCAAGGCACGUCGUCUUCGUUGUCAAUCAUAAUGACAACUUAACGCACACACUCUCAGCGAC-------- .............(((......(((.....(((((((.(((............))))))))))..)))(((((((.......)))))))..................)))..-------- ( -24.30) >DroEre_CAF1 317007 112 + 1 CAUUAACCAUCUCCGCACAAUUGAAUUAAUCGAUGUGGCCUAUUAGAAAAGCAAGGCACGUCGACUUUGUUGUCAAUCAUAAUGACAACUUAACGCACACACUCUCAGCGAC-------- .............(((.............((((((((.(((............)))))))))))....(((((((.......)))))))..................)))..-------- ( -25.40) >DroYak_CAF1 301551 112 + 1 CAUUAACUAUGUCCGCACAAUUGAAUUAAUCGACGUGGCCUAUUAGAAAACCAAGGCACGUCGACUUUGUUGUCAAUCAUAAUGACAACUUAACGCACACACUCUCAGCGAC-------- .............(((.............((((((((.(((............)))))))))))....(((((((.......)))))))..................)))..-------- ( -27.20) >DroAna_CAF1 462938 120 + 1 CAUUAACUGUCUCCGCACAAUUGAAUUAAUCGAUGCGGCCCAUUACGAAACCGAGGCACGUCGACUUUGUUGUCAAUCAUAAUGACAACUUAACGCACACUCACACACACACACGUGCAG ..............((((...........((((((..((((...........).))).))))))....(((((((.......))))))).........................)))).. ( -24.40) >consensus CAUUAACCAUCUCCGCACAAUUGAAUUAAUCGAUGUGGCCUAUUAGAAAACCAAGGCACGUCGACUUCGUUGUCAAUCAUAAUGACAACUUAACGCACACACUCUCAGCGAC________ ..............((.............((((((((.(((............)))))))))))....(((((((.......))))))).....))........................ (-21.41 = -21.93 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:08 2006