| Sequence ID | X_DroMel_CAF1 |
|---|---|
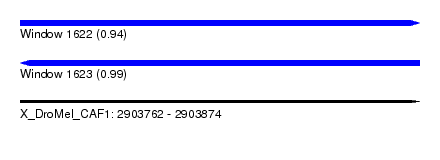
| Location | 2,903,762 – 2,903,874 |
| Length | 112 |
| Max. P | 0.990571 |

| Location | 2,903,762 – 2,903,874 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.33 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2903762 112 + 22224390 AAAACACAGAAAACAGCAGAA--CUGAAAAGCAGCAAACAGUUGCAAACUGGUAUGCCCCACGUUGCCAAUGUUGCUGCCGCUCCUCGCUCGCCCUGCCCUGCUGUCCCCACCA ............(((((((..--.......(((((((.((...((((..(((......)))..))))...))))))))).((.....))..........)))))))........ ( -30.50) >DroSec_CAF1 290488 91 + 1 A---------AAACAGUAGAA--CUGAAAAGCAGCAAACAGUUGCAAACUGGUAUGCCCCACGUUGCCGAUGUUGCUGCCGCUCCUCGCA----------UGCUGCCCCC--CC .---------...(((((...--.......(((((((.((...((((..(((......)))..))))...))))))))).((.....)).----------))))).....--.. ( -24.90) >DroYak_CAF1 298986 97 + 1 AAAACACAGA----------AAACAGAAAAGCAGCAAACAGUUGCAAACUGGUAUGCCCCACGUUGCUGAUGUUGCUGCCGUUCUUCG----CCCUG-CAUGCUGCCCCC--CG ......(((.----------....((((..(((((((.((...((((..(((......)))..))))...)))))))))..))))...----..)))-............--.. ( -25.40) >consensus AAAACACAGAAAACAG_AGAA__CUGAAAAGCAGCAAACAGUUGCAAACUGGUAUGCCCCACGUUGCCGAUGUUGCUGCCGCUCCUCGC___CCCUG_C_UGCUGCCCCC__CA ..............................(((((((.((...((((..(((......)))..))))...)))))))))................................... (-19.33 = -19.33 + -0.00)


| Location | 2,903,762 – 2,903,874 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -25.15 |
| Energy contribution | -25.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2903762 112 - 22224390 UGGUGGGGACAGCAGGGCAGGGCGAGCGAGGAGCGGCAGCAACAUUGGCAACGUGGGGCAUACCAGUUUGCAACUGUUUGCUGCUUUUCAG--UUCUGCUGUUUUCUGUGUUUU ....(..(((((((((((...(....)(((((((((((..((((...((((..(((......)))..))))...))))))))))))))).)--))))))))))..)........ ( -47.80) >DroSec_CAF1 290488 91 - 1 GG--GGGGGCAGCA----------UGCGAGGAGCGGCAGCAACAUCGGCAACGUGGGGCAUACCAGUUUGCAACUGUUUGCUGCUUUUCAG--UUCUACUGUUU---------U ..--.(((((((..----------.(((((((((((((((..((.((....))))..))....((((.....))))..))))))))))).)--)....))))))---------) ( -32.80) >DroYak_CAF1 298986 97 - 1 CG--GGGGGCAGCAUG-CAGGG----CGAAGAACGGCAGCAACAUCAGCAACGUGGGGCAUACCAGUUUGCAACUGUUUGCUGCUUUUCUGUUU----------UCUGUGUUUU ((--(..(((.((...-....)----)..((((.((((((((((...((((..(((......)))..))))...)).)))))))).))))))).----------.)))...... ( -33.00) >consensus CG__GGGGGCAGCA_G_CAGGG___GCGAGGAGCGGCAGCAACAUCGGCAACGUGGGGCAUACCAGUUUGCAACUGUUUGCUGCUUUUCAG__UUCU_CUGUUUUCUGUGUUUU ...........................(((((((((((..((((...((((..(((......)))..))))...)))))))))))))))......................... (-25.15 = -25.27 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:07 2006