| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 705,932 – 706,032 |
| Length | 100 |
| Max. P | 0.658615 |

| Location | 705,932 – 706,032 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 705932 100 + 1281640 GGCAAAUAUCAUACCCACAGCUCAUUGCUCGCCCACAAUAUCAAUUAGUCCUACCGACUCACUGUUAGGUAUGGGUGAACAUUCUGCAAAUCAACAAAAU ...................((....((.(((((((...(((((((.((((.....))))....))).))))))))))).))....))............. ( -18.30) >DroPse_CAF1 15690 100 + 1 AGCGAACACUAUACCCACCGCCCAUUGCUCACCCACAACCUCCAUAAGUCCAACGGACUCUCUUUUGGGUCUGGGCGAGCACUCUGUGAACAGAAACAUU .(((..............)))....(((((.((((..((((.....(((((...))))).......)))).)))).))))).((((....))))...... ( -28.64) >DroSim_CAF1 6461 100 + 1 GGCAAAUAUCAUACCCACAGCUCACUGCUCGCCCACAAUAUCAAUUAGUCCUACCGACUCACUUUUAGGUAUGGGUGAACAUUCUGCAAACCAACAAAAU .(((...............((.....))(((((((...........((((.....)))).(((....))).)))))))......)))............. ( -16.90) >DroEre_CAF1 19207 100 + 1 GGCAAAUAUCAUACCCACAGCUCACUGCUCGCCCACAAUCUCAGUUAGUCCAACCGACUCUCUUUUAGGUAUGGGUGAACAUUCUGCAAAUCAACAAAAU .(((...............((.....))(((((((..((((.((..((((.....))))..))...)))).)))))))......)))............. ( -20.20) >DroYak_CAF1 9693 100 + 1 GACAAAUAUCAUACCUACAGCUCACUGCUCGCCCACAAUCUCAAUAAGCCCUACCGACUCUCUUUUAGGUAUGGGUGAACAUUCUGGAAAUCAACAAAAU .............((....((.....))(((((((................((((............))))))))))).......))............. ( -13.99) >DroPer_CAF1 15859 100 + 1 AGCGAACACUAUACCCACCGCCCAUUGCUCACCCACAACCUCCAUAAGUCCAACGGACUCUCUUUUGGGUCUGGGCGAGCACUCUGUGAACAGAAACAUU .(((..............)))....(((((.((((..((((.....(((((...))))).......)))).)))).))))).((((....))))...... ( -28.64) >consensus GGCAAAUAUCAUACCCACAGCUCACUGCUCGCCCACAAUCUCAAUAAGUCCAACCGACUCUCUUUUAGGUAUGGGUGAACAUUCUGCAAACCAACAAAAU .(((...............((.....))(((((((..((((.....((((.....)))).......)))).)))))))......)))............. (-13.73 = -13.90 + 0.17)

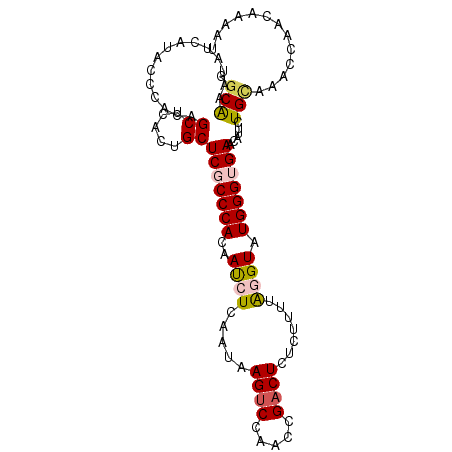
| Location | 705,932 – 706,032 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -22.88 |
| Energy contribution | -22.58 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 705932 100 - 1281640 AUUUUGUUGAUUUGCAGAAUGUUCACCCAUACCUAACAGUGAGUCGGUAGGACUAAUUGAUAUUGUGGGCGAGCAAUGAGCUGUGGGUAUGAUAUUUGCC ....((((((((..(((..(((((.((((((..((.((((.((((.....)))).)))).)).)))))).))))).....)))..))).)))))...... ( -28.60) >DroPse_CAF1 15690 100 - 1 AAUGUUUCUGUUCACAGAGUGCUCGCCCAGACCCAAAAGAGAGUCCGUUGGACUUAUGGAGGUUGUGGGUGAGCAAUGGGCGGUGGGUAUAGUGUUCGCU .....(((((....)))))(((((((((((((((....(.((((((...)))))).).).)))).))))))))))..(((((.((....)).)))))... ( -36.40) >DroSim_CAF1 6461 100 - 1 AUUUUGUUGGUUUGCAGAAUGUUCACCCAUACCUAAAAGUGAGUCGGUAGGACUAAUUGAUAUUGUGGGCGAGCAGUGAGCUGUGGGUAUGAUAUUUGCC ........((((..(....(((((.((((((......(((.((((.....)))).))).....)))))).))))))..))))...((((.......)))) ( -26.10) >DroEre_CAF1 19207 100 - 1 AUUUUGUUGAUUUGCAGAAUGUUCACCCAUACCUAAAAGAGAGUCGGUUGGACUAACUGAGAUUGUGGGCGAGCAGUGAGCUGUGGGUAUGAUAUUUGCC ....((((((((..(((..(((((.((((((.((...((..((((.....))))..)).))..)))))).))))).....)))..))).)))))...... ( -29.00) >DroYak_CAF1 9693 100 - 1 AUUUUGUUGAUUUCCAGAAUGUUCACCCAUACCUAAAAGAGAGUCGGUAGGGCUUAUUGAGAUUGUGGGCGAGCAGUGAGCUGUAGGUAUGAUAUUUGUC ((((((........)))))).......((((((((...(.....).....(((((((((...(((....))).))))))))).))))))))......... ( -23.40) >DroPer_CAF1 15859 100 - 1 AAUGUUUCUGUUCACAGAGUGCUCGCCCAGACCCAAAAGAGAGUCCGUUGGACUUAUGGAGGUUGUGGGUGAGCAAUGGGCGGUGGGUAUAGUGUUCGCU .....(((((....)))))(((((((((((((((....(.((((((...)))))).).).)))).))))))))))..(((((.((....)).)))))... ( -36.40) >consensus AUUUUGUUGAUUUGCAGAAUGUUCACCCAUACCUAAAAGAGAGUCGGUAGGACUAAUUGAGAUUGUGGGCGAGCAAUGAGCUGUGGGUAUGAUAUUUGCC .........(..(((((..(((((.((((((..........((((.....)))).........)))))).))))).....)))))..)............ (-22.88 = -22.58 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:28 2006