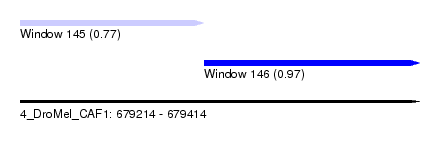
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 679,214 – 679,414 |
| Length | 200 |
| Max. P | 0.974600 |

| Location | 679,214 – 679,306 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 93.48 |
| Mean single sequence MFE | -16.81 |
| Consensus MFE | -14.77 |
| Energy contribution | -14.99 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 679214 92 + 1281640 CAUACAUAUGGGUAUAUAAGAGUGGGAGUCAUUUGACAGGAGUUAAACAUGUAUACUUAGUAUGUACAUAAAGAAUUUUAAAAAUGAAAUAU ..((((((((((((((((.........(((....))).(........).)))))))))).))))))........((((((....)))))).. ( -15.30) >DroSec_CAF1 12834 92 + 1 CAUACAUAUGAGUAUAUAAGAGUGGCAGUAAUUUGACAGGAGUUACACAUAUAUACUUAGUAUGUACAUAUAAAAUUAUAAAACUGAAAUAU ..((((((((((((((((.(.(((((...............))))).).)))))))))).)))))).......................... ( -17.56) >DroSim_CAF1 11083 92 + 1 CAUACAUAUGAGUAUAUAAGAGUGGCAGUAAUUUGACAGGAGUUACACAUAUAUACUUAGUAUGUACAUAUAAAAUUAUAAAACUGAAAUAU ..((((((((((((((((.(.(((((...............))))).).)))))))))).)))))).......................... ( -17.56) >consensus CAUACAUAUGAGUAUAUAAGAGUGGCAGUAAUUUGACAGGAGUUACACAUAUAUACUUAGUAUGUACAUAUAAAAUUAUAAAACUGAAAUAU ..((((((((((((((((.(.(((((...............))))).).)))))))))).)))))).......................... (-14.77 = -14.99 + 0.23)

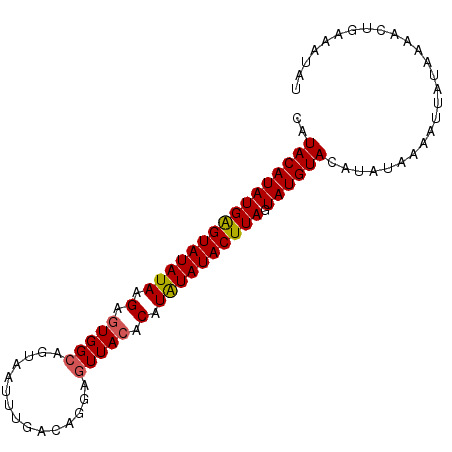
| Location | 679,306 – 679,414 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.36 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -9.96 |
| Energy contribution | -9.56 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 679306 108 + 1281640 GCAUCGCGAUCGCUGACCUACCAGGUCUGCUGCCCAACCGGCAGAAUCCCCAC--CCACAUUAACGACAAAUUCCUGUGUGAAAAAC--------GAAUGCAUGUCAAAAAAUAAAAC ((((.(((.(((..((((.....))))..(((((.....))))).........--.(((((....((.....))..))))).....)--------)).)))))))............. ( -23.30) >DroSec_CAF1 12926 86 + 1 G-----------------------GCCUGCUGCCCAACCGGCAGUAUCCCCACACGCACAUUAACGACAAAUUCACGUGUGAAAAAC--------GAAUGCAUGCCAAAAAA-AUAAA (-----------------------((.(((((((.....)))))))....((((((...................))))))......--------........)))......-..... ( -20.81) >DroSim_CAF1 11175 109 + 1 GCAUCGCGAUCGCUGCCCUAACAGGCCUGCUGCCCAACCGGCAGUAUCCCCACACGCACAUUGACGACAAAUUCACGUGUGAAAAAC--------GAAUGCAUGCCAAAAAA-AUAAA ((((.(((.(((..(((......))).(((((((.....)))))))....((((((....(((....))).....)))))).....)--------)).))))))).......-..... ( -29.80) >DroEre_CAF1 25523 116 + 1 GUAUUGCGAUCGCUGCACUACCAGUACUGCUGUCCAACCGGCAGUUCCCCCAGGCGCACAUAAACGAAAAAUUCACGUGUGACAAACAUACAUACAUAUGUAUGUCAAAAU--AUAAA ((((((((.((((((......))))(((((((......))))))).......)))))......................(((((.(((((......))))).))))).)))--))... ( -26.00) >DroYak_CAF1 14912 107 + 1 AUAUGGCGAUCGGUGCACUACCAGGGCUACUGCCAAACCGGUAGUUUC-CCAGGCGCAUAUUAACGAAAAAUUCACGUGUGGAAAAC--------AUUUGUAUAUCAAAAA--UUAAA (((((((....((((...)))).(((..((((((.....))))))..)-))..)).)))))....((......((.((((.....))--------)).))....)).....--..... ( -24.70) >consensus GCAUCGCGAUCGCUGCACUACCAGGCCUGCUGCCCAACCGGCAGUAUCCCCACACGCACAUUAACGACAAAUUCACGUGUGAAAAAC________GAAUGCAUGUCAAAAAA_AUAAA ............................((((((.....))))))...........(((((....((.....))..)))))..................................... ( -9.96 = -9.56 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:26 2006