| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 669,650 – 669,785 |
| Length | 135 |
| Max. P | 0.723058 |

| Location | 669,650 – 669,757 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.51 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -18.68 |
| Energy contribution | -18.10 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
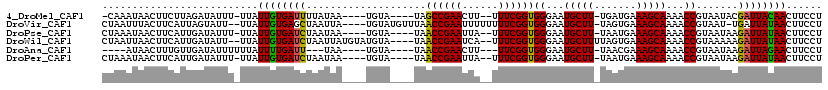
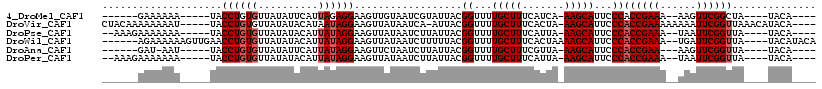
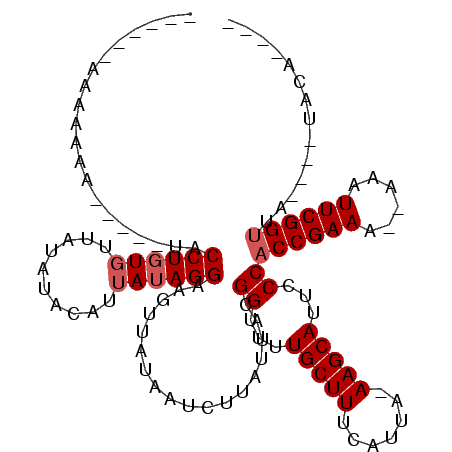
>4_DroMel_CAF1 669650 107 + 1281640 -CAAAUAACUUCUUAGAUAUUU-UUAUUGUGAUUUUAUAA----UGUA----UAGCCGAACUU--UUUCGGUGGGAAUGCUU-UGAUGAAAGCAAAACCGUAAUACGAUUACAACUUCCU -.(((((.((....)).)))))-...((((((((.(((..----....----..((((((...--.))))))((...(((((-(....))))))...))...))).))))))))...... ( -21.60) >DroVir_CAF1 6853 112 + 1 CUAAUUUACUUCAUUAGUAUU--UUAUUGUGAGCUAAUUA----UGUAUGUUUAACCGAAUUUUUUUUCGGUGGGAAUGCUU-UAGUGAAAGCAAAACCGUAAU-UGAUUAUAACUUCCU .(((..((((.....))))..--)))((((((..((((((----(.........((((((......))))))((...(((((-(....))))))...)))))))-)).))))))...... ( -24.10) >DroPse_CAF1 9023 108 + 1 CUAAAUAACUUCAUUGAUAUUU-UUAUUGUGAUCUAAUAA----UGUA----UAACCGAAUUA--UUUCGGUGGGAAUGCUU-UAAUGAAAGCAAAACCGUAAUAAGAUUAUAACUUCCU ......................-...(((((((((.....----....----..((((((...--.))))))((...(((((-(....))))))...))......)))))))))...... ( -22.40) >DroWil_CAF1 6423 112 + 1 CUAAUUAACUUCAUUGAUAUU--UUAUUGUGAUCUAAUUAUGUAUGUA----UAACCGAAUCA--UUUCGGUGGGAAUGCUUUUAGUGAAAGCAAAACCGUAAAAAGAUUAUAACUUCCU .....................--...(((((((((..((((.......----..((((((...--.))))))((...(((((((...)))))))...))))))..)))))))))...... ( -23.30) >DroAna_CAF1 1585406 101 + 1 ----AUAACUUUGUUGAUAUUUUUUAUUUUGAUU---UAA----UGUA----UAACCGAACUU---UUCGGUGGGAAUGCUU-UAACGAAAGCAAAACCGUAAUAAGAUUAGAACUUCCU ----......................((((((((---(..----....----..(((((....---.)))))((...(((((-(....))))))...))......)))))))))...... ( -17.60) >DroPer_CAF1 15582 108 + 1 CUAAAUAACUUCAUUGAUAUUU-UUAUUGUGAUCUAAUAA----UGUA----UAACCGAAUUA--UUUCGGUGGGAAUGCUU-UAAUGAAAGCAAAACCGUAAUAAGAUUAUAACUUCCU ......................-...(((((((((.....----....----..((((((...--.))))))((...(((((-(....))))))...))......)))))))))...... ( -22.40) >consensus CUAAAUAACUUCAUUGAUAUUU_UUAUUGUGAUCUAAUAA____UGUA____UAACCGAAUUA__UUUCGGUGGGAAUGCUU_UAAUGAAAGCAAAACCGUAAUAAGAUUAUAACUUCCU ..........................((((((((....................((((((......))))))((...(((((.......)))))...)).......))))))))...... (-18.68 = -18.10 + -0.58)
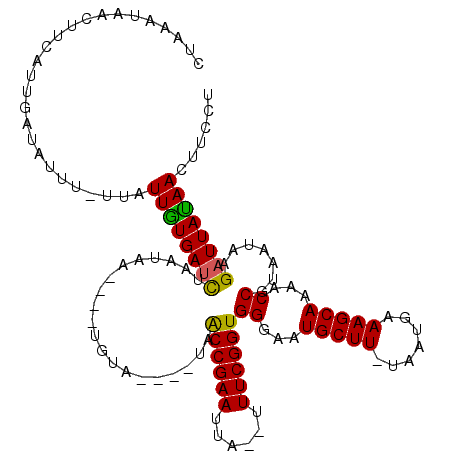
| Location | 669,688 – 669,785 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -18.73 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.72 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 669688 97 - 1281640 ------GAAAAAA-----UACCUGUGUUAUAUUCAUUAGAGGAAGUUGUAAUCGUAUUACGGUUUUGCUUUCAUCA-AAGCAUUCCCACCGAAA--AAGUUCGGCUA----UACA---- ------.......-----.....((((...........((((((((...(((((.....)))))..)))))).)).-...........(((((.--...)))))..)----))).---- ( -17.20) >DroVir_CAF1 6891 108 - 1 CUACAAAAAAAAU-----UACCUGUGUUAUAUACAUAAUAGGAAGUUAUAAUCA-AUUACGGUUUUGCUUUCACUA-AAGCAUUCCCACCGAAAAAAAAUUCGGUUAAACAUACA---- .............-----..(((((............)))))..(((.......-.....((...((((((....)-)))))...))((((((......))))))..))).....---- ( -18.10) >DroPse_CAF1 9062 101 - 1 --AAAGAAAAAAA-----UACCUGUGUUAUAUACAUUAUAGGAAGUUAUAAUCUUAUUACGGUUUUGCUUUCAUUA-AAGCAUUCCCACCGAAA--UAAUUCGGUUA----UACA---- --...........-----..((((((..........))))))..((.(((((((((((.((((..((((((....)-))))).....)))).))--)))...)))))----))).---- ( -19.50) >DroWil_CAF1 6461 107 - 1 ------AGAAAAAAGUUGAACCUGUGUUAUAUACAUUAUAGGAAGUUAUAAUCUUUUUACGGUUUUGCUUUCACUAAAAGCAUUCCCACCGAAA--UGAUUCGGUUA----UACAUACA ------.(.((((((.(((.((((((..........))))))...)))....)))))).)((...((((((.....))))))...))((((((.--...))))))..----........ ( -20.40) >DroAna_CAF1 1585439 95 - 1 ------GAU-AAU-----UACCUGUGUUAUAUUCAUUAUAGGAAGUUCUAAUCUUAUUACGGUUUUGCUUUCGUUA-AAGCAUUCCCACCGAA---AAGUUCGGUUA----UACA---- ------(((-(((-----(.((((((..........)))))).))))...))).......((...((((((....)-)))))...))(((((.---....)))))..----....---- ( -17.70) >DroPer_CAF1 15621 101 - 1 --AAAGAAAAAAA-----UACCUGUGUUAUAUACAUUAUAGGAAGUUAUAAUCUUAUUACGGUUUUGCUUUCAUUA-AAGCAUUCCCACCGAAA--UAAUUCGGUUA----UACA---- --...........-----..((((((..........))))))..((.(((((((((((.((((..((((((....)-))))).....)))).))--)))...)))))----))).---- ( -19.50) >consensus ______AAAAAAA_____UACCUGUGUUAUAUACAUUAUAGGAAGUUAUAAUCUUAUUACGGUUUUGCUUUCAUUA_AAGCAUUCCCACCGAAA__AAAUUCGGUUA____UACA____ ....................((((((..........))))))..................((...(((((.......)))))...))((((((......)))))).............. (-15.22 = -15.72 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:19 2006