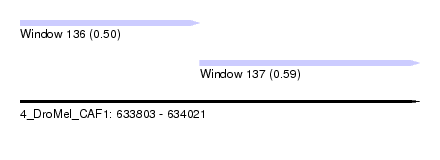
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 633,803 – 634,021 |
| Length | 218 |
| Max. P | 0.594021 |

| Location | 633,803 – 633,901 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -18.52 |
| Energy contribution | -19.47 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 633803 98 + 1281640 UCCUAUAAAGUUU----------------------CUUUAUGUACCGAUACAAAAUAUCUCCUUCAUUUAAACUUAUUUUUAGUGGGUGGAGGAGUCAUUUACCGACUUCGUUAAGGGCA (((((..(((((.----------------------.....((((....))))((((..((((((((((...(((.......))).))))))))))..))))...)))))..)..)))).. ( -20.90) >DroSec_CAF1 9773 117 + 1 UCCUAUAAAGUUAGAUUUGGCUUAGAGAAGUUACAUUUUAUUAACCGGU--AAGAUAUCUCCUUCAUAUUAACUCAUU-UUAGUGGGUGGAGGAGUCAUUUACCGACUUCGUUAAGGGCA (((((..(((((((((.((((((....)))))).))))........(((--(((....(((((((((....(((....-..)))..)))))))))...)))))))))))..)..)))).. ( -29.20) >DroSim_CAF1 15844 117 + 1 UCCUAUAAAGUUAGAUUUGGCUUAGAGAAGUUACAUUUUAUUAACCGGU--AAGAUAUCUCCUUCAUAUUAACUUAUU-UUAGUGGGUGGAGGAAUCAUUUACCGACUUCGUUAAGGGCA (((((..(((((((((.((((((....)))))).))))........(((--(((.....((((((((....(((....-..)))..))))))))....)))))))))))..)..)))).. ( -26.30) >consensus UCCUAUAAAGUUAGAUUUGGCUUAGAGAAGUUACAUUUUAUUAACCGGU__AAGAUAUCUCCUUCAUAUUAACUUAUU_UUAGUGGGUGGAGGAGUCAUUUACCGACUUCGUUAAGGGCA (((((..((((......((((((....))))))............((((...((((..(((((((((....(((.......)))..)))))))))..))))))))))))..)..)))).. (-18.52 = -19.47 + 0.95)

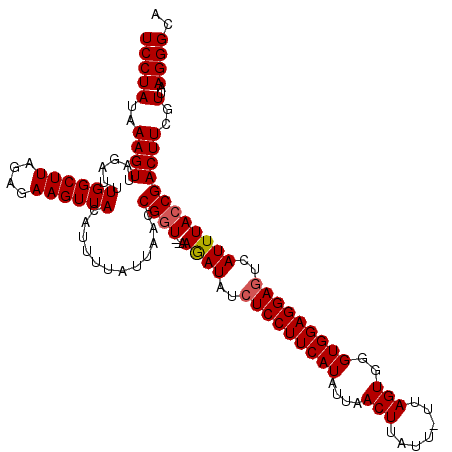
| Location | 633,901 – 634,021 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.66 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 633901 120 + 1281640 UUAACUGGAGGAGCUAUGUGGUUUGCGAUGUGGCAUAUCACAACGUCAAUAACUGGUUGUGGUCGCCGUUUAUCGAUCGUGGAUCAGCAAACCGACUUUACAUUGAAAUUCAGUUCACAA ..(((((((....(.(((((((((((((((((((..((((((((...........)))))))).))))..))))((((...)))).)))))))......)))).)...)))))))..... ( -35.60) >DroGri_CAF1 4703 120 + 1 UUAAUUGGCGUAGUUAUGUGGUUUGCGAUGUUGCCUAUCACAAUGUUAACAACUGGCUCUGGUCCCCAUUCAUUGACCGUGGACCUGCUAACCGUCUUUAUAUAGAAAUACAAUUUACAA ......((((..((((((((((..(((....)))..))))))....))))...((((...(((((.(...........).))))).))))..))))........................ ( -24.40) >DroSec_CAF1 9890 120 + 1 UAAACUGGAGAAGCUAUGUGGUUUGCGAUGUGGCAUAUCACAACGUCAAUAACUGGUUGUGGUCGCCCUUUAUCGAUCGUGGAUCAGCAAACCGACUUUACAUUGAGAUUCAGUUCACAA ..(((((((.........((((((((((((.(((..((((((((...........)))))))).)))...))))((((...)))).)))))))).(((......))).)))))))..... ( -35.50) >DroSim_CAF1 15961 120 + 1 UAAACUGGAGAAGCUAUGUGGUUUGCGAUGUGGCAUAUCACAACGUCAAUAACUGGUUGUGGUCGCCGUUUAUCGAUCGUGGAUCAGCAAACCGACUUUACAUUGAGAUUCAGUUCACAA ..(((((((.........((((((((((((((((..((((((((...........)))))))).))))..))))((((...)))).)))))))).(((......))).)))))))..... ( -35.80) >DroYak_CAF1 1634 120 + 1 UAAACUGGAGAAGCUAUGUGGUGUGCGAUGUGGCAUAUCACAACGUCAAUAACUGGUUGUGGUCGCCGUUUAUUGAUCGUGGAUCAGCAAACCGACUUUAUAUUGAAAUUCAGUUCACAA ..(((((((.......((((((((((......))))))))))...((((((((.....))(((((..((((.((((((...)))))).)))))))))...))))))..)))))))..... ( -34.70) >consensus UAAACUGGAGAAGCUAUGUGGUUUGCGAUGUGGCAUAUCACAACGUCAAUAACUGGUUGUGGUCGCCGUUUAUCGAUCGUGGAUCAGCAAACCGACUUUACAUUGAAAUUCAGUUCACAA ..(((((((....(.(((((((((((((((.(((..((((((((...........)))))))).)))...))))((((...)))).)))))))......)))).)...)))))))..... (-25.94 = -26.66 + 0.72)

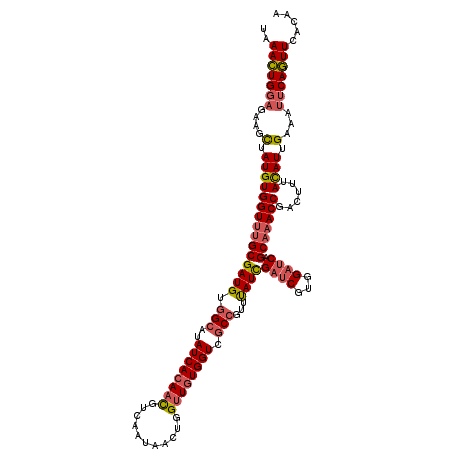
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:13 2006