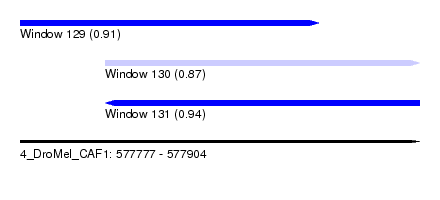
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 577,777 – 577,904 |
| Length | 127 |
| Max. P | 0.938120 |

| Location | 577,777 – 577,872 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -15.50 |
| Energy contribution | -16.81 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 577777 95 + 1281640 AAUUGAAGC-------------ACUGCUCCUUAACUAGCGUUCGCAGUAUGAUGCACAUUUUGCAUAAUUUCAAUUUCGAAUGUACAUGCAGACAUAUGUAUGUGCAC ((((((((.-------------(((((.(((.....)).)...)))))...(((((.....)))))..)))))))).....((((((((((......)))))))))). ( -25.80) >DroSec_CAF1 8096 95 + 1 AAUUGAAGC-------------ACUGCUCCUUAACUGGCGUUCGCAGUAUGAUGCACAUUUUGCAUAAUUUCAAUUUCAAAUGUAUAUGCAGACAUAUGUAUGUGCAC ((((((((.-------------(((((.((......)).....)))))...(((((.....)))))..)))))))).....((((((((((......)))))))))). ( -25.00) >DroSim_CAF1 7963 95 + 1 AAUUGAAGC-------------ACUGCUCCUUAACUGGCGUUCGCAGUAUGAUGCACAUUUUGCAUAAUUUCAAUUUCAAAUGUACAUGCAGACAUAUGUAUGUGCAC ((((((((.-------------(((((.((......)).....)))))...(((((.....)))))..)))))))).....((((((((((......)))))))))). ( -26.90) >DroEre_CAF1 9688 95 + 1 CAUUGAAGC-------------ACUGCUCCUUAACUGGCGUUCGCAGUAUGAUGCACAUUUUGCAUAAUUUCAAUUUCGAAUGUACAUGCAGACAUAUGUAUGUGCAC .(((((((.-------------(((((.((......)).....)))))...(((((.....)))))..)))))))......((((((((((......)))))))))). ( -26.30) >DroWil_CAF1 8537 96 + 1 --UUCACACUUUGGCCGUCCAAAUUUAUUUUGUAUUUGUUUUUGCGUUCUAGUACACAUUCUGCAUAAUUUCAAUUUCGUAUGUAU----------ACAUAUGUGCAU --.....(((..(..(((..((((..((.....))..))))..)))..).)))........((((............((((((...----------.)))))))))). ( -10.90) >DroYak_CAF1 8743 95 + 1 GAUUGAAGC-------------ACUGCUCCUUAACUGCCGUUCGAAGUAUGAUGCACAUUUUGCAUAAUUUGAAUUUCGAAUGUACAUGCAGACAUAUGUAUGUGCAC .......((-------------(((((.......((((((((((((((...(((((.....))))).......)))))))))).....))))......))).)))).. ( -24.72) >consensus AAUUGAAGC_____________ACUGCUCCUUAACUGGCGUUCGCAGUAUGAUGCACAUUUUGCAUAAUUUCAAUUUCGAAUGUACAUGCAGACAUAUGUAUGUGCAC ..((((((..............(((((................)))))...(((((.....)))))........)))))).((((((((((......)))))))))). (-15.50 = -16.81 + 1.31)

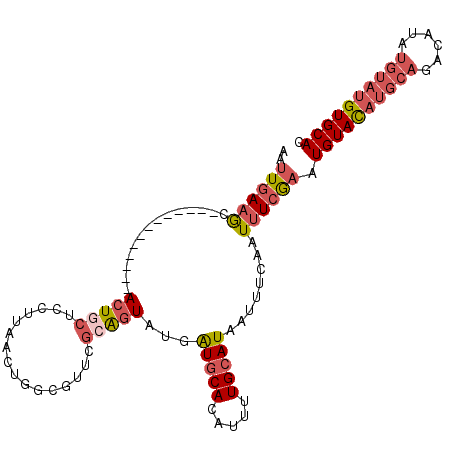
| Location | 577,804 – 577,904 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -15.20 |
| Energy contribution | -16.28 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 577804 100 + 1281640 UUCGCAGUAUGAUGCACAUUUUGCAUAAUUUCAAUUUCGAAUGUACAUGCAGACAUAUGUAUGUGCACACAUGUC---GUGUAUGAAUACACACACACC-----CGAA ...((((.(((.....))).))))...........((((..((((((((((......))))))))))....(((.---(((((....))))))))....-----)))) ( -25.30) >DroSec_CAF1 8123 100 + 1 UUCGCAGUAUGAUGCACAUUUUGCAUAAUUUCAAUUUCAAAUGUAUAUGCAGACAUAUGUAUGUGCACACAUGUC---GUGUAUGAAUACACACACACC-----CGAA .(((..((((((((((.....)))))...............((((((((((......))))))))))..)))).)---(((((....))))).......-----))). ( -22.30) >DroSim_CAF1 7990 100 + 1 UUCGCAGUAUGAUGCACAUUUUGCAUAAUUUCAAUUUCAAAUGUACAUGCAGACAUAUGUAUGUGCACACAUGUC---GUGUAUGAAUACACACACACC-----CGAA .(((..((((((((((.....)))))...............((((((((((......))))))))))..)))).)---(((((....))))).......-----))). ( -24.20) >DroEre_CAF1 9715 100 + 1 UUCGCAGUAUGAUGCACAUUUUGCAUAAUUUCAAUUUCGAAUGUACAUGCAGACAUAUGUAUGUGCACACAUGUC---GGGUAUGAAUACACACACAGC-----CGCA ...((.((.(((((((.....)))))...((((..(((((.((((((((((......))))))))))......))---)))..))))........))))-----.)). ( -24.40) >DroWil_CAF1 8575 88 + 1 UUUGCGUUCUAGUACACAUUCUGCAUAAUUUCAAUUUCGUAUGUAU----------ACAUAUGUGCAUACUAGUAUCCGCCCCCGCACUCGCACACAC---------- ...(((..(((((((((((..((((((............)))))).----------....)))))..))))))....)))....((....))......---------- ( -16.00) >DroYak_CAF1 8770 105 + 1 UUCGAAGUAUGAUGCACAUUUUGCAUAAUUUGAAUUUCGAAUGUACAUGCAGACAUAUGUAUGUGCACACAUGUC---GUGUAUGAAUACACACACCACAAACACGCA ((((((((...(((((.....))))).......))))))))((((((((((......))))))))))....(((.---((((.((....)).)))).)))........ ( -28.40) >consensus UUCGCAGUAUGAUGCACAUUUUGCAUAAUUUCAAUUUCGAAUGUACAUGCAGACAUAUGUAUGUGCACACAUGUC___GUGUAUGAAUACACACACACC_____CGAA ...........(((((.....)))))...............((((((((((......))))))))))...........(((((....)))))................ (-15.20 = -16.28 + 1.08)

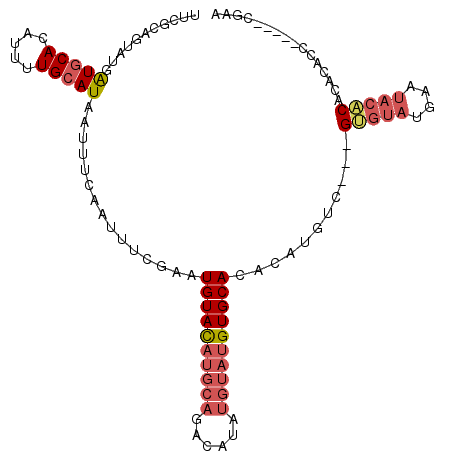
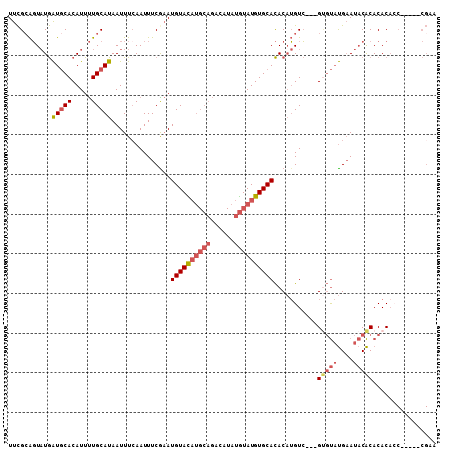
| Location | 577,804 – 577,904 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -17.78 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 577804 100 - 1281640 UUCG-----GGUGUGUGUGUAUUCAUACAC---GACAUGUGUGCACAUACAUAUGUCUGCAUGUACAUUCGAAAUUGAAAUUAUGCAAAAUGUGCAUCAUACUGCGAA .(((-----((((((((..((((.......---((((((((((....))))))))))((((((....((((....))))..)))))).))))..)).)))))).))). ( -31.40) >DroSec_CAF1 8123 100 - 1 UUCG-----GGUGUGUGUGUAUUCAUACAC---GACAUGUGUGCACAUACAUAUGUCUGCAUAUACAUUUGAAAUUGAAAUUAUGCAAAAUGUGCAUCAUACUGCGAA ((((-----((((((((((((.........---((((((((((....))))))))))))))))))))))))))..........((((..(((.....)))..)))).. ( -30.80) >DroSim_CAF1 7990 100 - 1 UUCG-----GGUGUGUGUGUAUUCAUACAC---GACAUGUGUGCACAUACAUAUGUCUGCAUGUACAUUUGAAAUUGAAAUUAUGCAAAAUGUGCAUCAUACUGCGAA .(((-----((((((((((....)))))))---((((((((((....))))))))))((.((((((((((...((.......))...)))))))))))).))).))). ( -32.40) >DroEre_CAF1 9715 100 - 1 UGCG-----GCUGUGUGUGUAUUCAUACCC---GACAUGUGUGCACAUACAUAUGUCUGCAUGUACAUUCGAAAUUGAAAUUAUGCAAAAUGUGCAUCAUACUGCGAA .(((-----(.((((((..((((.......---((((((((((....))))))))))((((((....((((....))))..)))))).))))..)).))))))))... ( -31.10) >DroWil_CAF1 8575 88 - 1 ----------GUGUGUGCGAGUGCGGGGGCGGAUACUAGUAUGCACAUAUGU----------AUACAUACGAAAUUGAAAUUAUGCAGAAUGUGUACUAGAACGCAAA ----------.....((((..(((....)))....((((((((((....(((----------(((((........))....))))))...))))))))))..)))).. ( -21.50) >DroYak_CAF1 8770 105 - 1 UGCGUGUUUGUGGUGUGUGUAUUCAUACAC---GACAUGUGUGCACAUACAUAUGUCUGCAUGUACAUUCGAAAUUCAAAUUAUGCAAAAUGUGCAUCAUACUUCGAA (((((((.....(((((((....)))))))---((((((((((....)))))))))).)))))))..((((((.........(((((.....))))).....)))))) ( -32.14) >consensus UUCG_____GGUGUGUGUGUAUUCAUACAC___GACAUGUGUGCACAUACAUAUGUCUGCAUGUACAUUCGAAAUUGAAAUUAUGCAAAAUGUGCAUCAUACUGCGAA ............(((((((....)))))))...((((((((((....))))))))))...(((((((((....((.......))....)))))))))........... (-17.78 = -18.62 + 0.84)

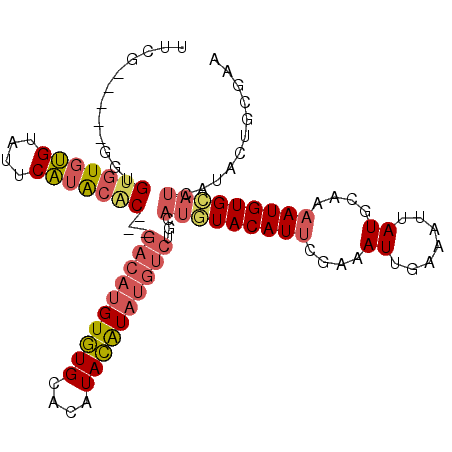
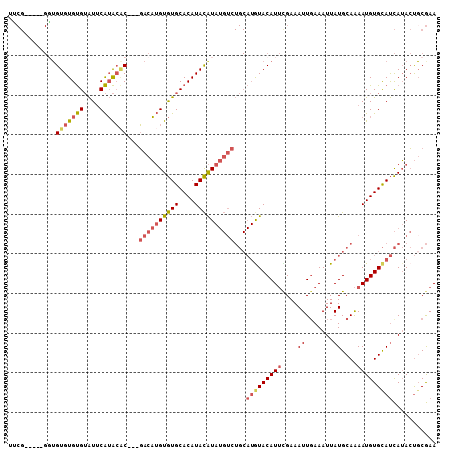
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:05 2006