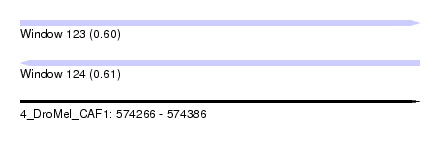
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 574,266 – 574,386 |
| Length | 120 |
| Max. P | 0.609530 |

| Location | 574,266 – 574,386 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.04 |
| Mean single sequence MFE | -17.58 |
| Consensus MFE | -10.43 |
| Energy contribution | -10.21 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 574266 120 + 1281640 UAUUACCAUAAUCGUCGUCAUCUGCCGAACACAAAUAUUAUUUACCAUCUUAUUCUCUCCUUAUUAUUACACUAUCUGUGCGCUAUUUAAACUUUAUAUAGCCCGGUAAAUGACGAUGUU ............(((((((((.(((((..........................................(((.....))).(((((.((.....)).))))).))))).))))))))).. ( -25.50) >DroSec_CAF1 4779 97 + 1 --UUACAUUAAUUGUCGUCAUCUGCCGAACACAAACAUUAUUUACCGUCUUAUUCUCUC---------------------CGCGAUUUAAACUUUAUACGGCUCGGUAUAUGACUAUGUU --..............(((((.((((((....(((.....))).((((...........---------------------.................)))).)))))).)))))...... ( -12.99) >DroSim_CAF1 4746 94 + 1 --UUACAUUAAUCGUCGUCAUCUGCCGAACACAAACAUUAUUUACCGUCUUA---UCUC---------------------CGCGAUUUAAACUUUAUACGGCUCGGUAUAUGACUAUGUU --..........(((.(((((.((((((....(((.....))).((((...(---((..---------------------...)))...........)))).)))))).))))).))).. ( -14.24) >consensus __UUACAUUAAUCGUCGUCAUCUGCCGAACACAAACAUUAUUUACCGUCUUAUUCUCUC_____________________CGCGAUUUAAACUUUAUACGGCUCGGUAUAUGACUAUGUU ............(((.(((((.(((((............................................................................))))).))))).))).. (-10.43 = -10.21 + -0.22)

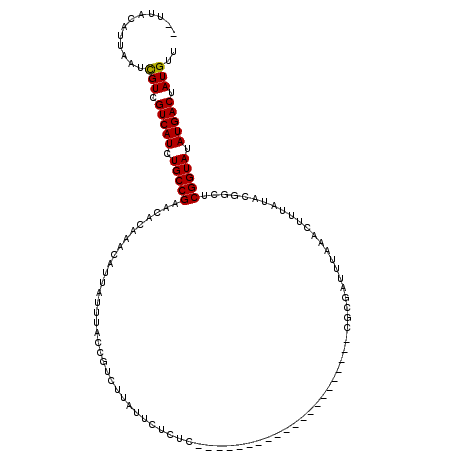
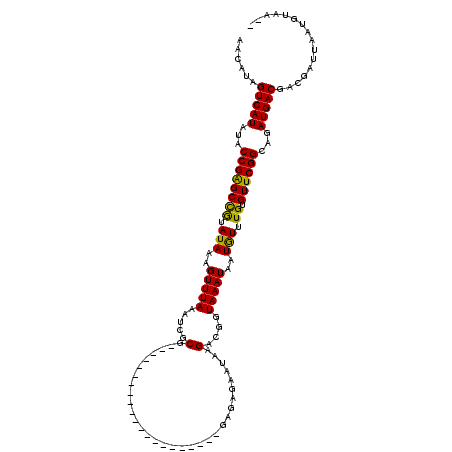
| Location | 574,266 – 574,386 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.04 |
| Mean single sequence MFE | -21.81 |
| Consensus MFE | -15.40 |
| Energy contribution | -14.52 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 574266 120 - 1281640 AACAUCGUCAUUUACCGGGCUAUAUAAAGUUUAAAUAGCGCACAGAUAGUGUAAUAAUAAGGAGAGAAUAAGAUGGUAAAUAAUAUUUGUGUUCGGCAGAUGACGACGAUUAUGGUAAUA ..(.(((((((((.(((((((((.((.....)).)))))(((((((((.(((...........................))).))))))))))))).))))))))).)............ ( -27.63) >DroSec_CAF1 4779 97 - 1 AACAUAGUCAUAUACCGAGCCGUAUAAAGUUUAAAUCGCG---------------------GAGAGAAUAAGACGGUAAAUAAUGUUUGUGUUCGGCAGAUGACGACAAUUAAUGUAA-- ......(((((...((((((((.(((..((((..(((((.---------------------..........).))))))))..))).)).))))))...)))))..............-- ( -18.00) >DroSim_CAF1 4746 94 - 1 AACAUAGUCAUAUACCGAGCCGUAUAAAGUUUAAAUCGCG---------------------GAGA---UAAGACGGUAAAUAAUGUUUGUGUUCGGCAGAUGACGACGAUUAAUGUAA-- .((((((((.(((((......)))))..(((...(((((.---------------------(((.---..(((((........)))))...))).)).)))...))))))).))))..-- ( -19.80) >consensus AACAUAGUCAUAUACCGAGCCGUAUAAAGUUUAAAUCGCG_____________________GAGAGAAUAAGACGGUAAAUAAUGUUUGUGUUCGGCAGAUGACGACGAUUAAUGUAA__ ......(((((...((((((((.(((..(((((.....(................................)....)))))..))).)).))))))...)))))................ (-15.40 = -14.52 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:48 2006