| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 566,028 – 566,142 |
| Length | 114 |
| Max. P | 0.517082 |

| Location | 566,028 – 566,142 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.53 |
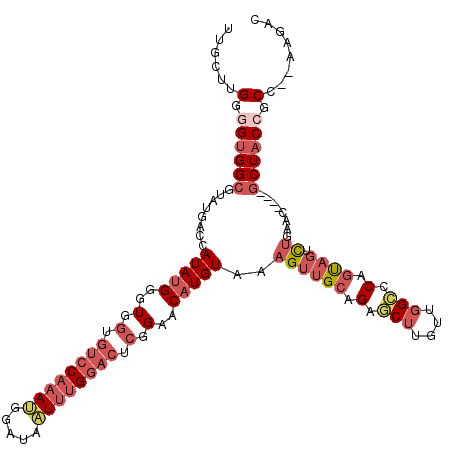
| Mean single sequence MFE | -43.42 |
| Consensus MFE | -21.44 |
| Energy contribution | -23.63 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 566028 114 + 1281640 UUGCUUGGGGUGGCGUAUGACCAUAUGGGUGGUGUCCAAAUGGAUAAUUUGGACUCGGAACAUGUAAAGUUGCAGAGCUUGUAGGCACAGCAGUCUGAAC----GCUACCGCC--CAGAC ...((.((((((((((..(((.(((((..(.(.((((((((.....)))))))).).)..))))).....(((.(.(((....))).).))))))...))----)))))).))--.)).. ( -42.60) >DroSec_CAF1 13267 114 + 1 UUGCUUGGGGUGGCGUAUGACCAUAUGGGUGGUGUCCAAAUGGAUAAUUUGGACUCGGAACAUGUAAAGUUGCAGAGCUUGUUGGCCCAGUAGUCUGAAC----GCUACCGCC--AAGAC ...(((((((((((((..(((....(((((.(.((((((((.....)))))))).)....((....(((((....)))))..)))))))...)))...))----)))))).))--))).. ( -46.20) >DroSim_CAF1 16204 114 + 1 UUGCUUGGGGUGGCGUAUGACCAUAUGGGUGGUGUCCAAAUGGAUAAUUUGGACUCGGAACAUGUAAAGUUGCAGAGCUUGUUGGCCCAGUAGUCUGAAC----GCUACCGCC--AAGAC ...(((((((((((((..(((....(((((.(.((((((((.....)))))))).)....((....(((((....)))))..)))))))...)))...))----)))))).))--))).. ( -46.20) >DroEre_CAF1 19009 114 + 1 UUGCUUGGUGUGGCGUAUGACCAUAUGGGUGGUGUCCAAAGGGAUAAUUUGGACUCGGAACAUGUAAAGUUGCAGUGCUAGUUGGCCCAGUAGUCUGAAC----GCUACUGCC--CAGAC ...((.((((((((((..(((....(((((.(.(((((((.......))))))).)((.((.((((....)))))).)).....)))))...)))...))----))))).)))--.)).. ( -37.50) >DroWil_CAF1 12635 117 + 1 UAGCCUGCGGCGGGGUGUGCCCAUAAGGAUG---AGCAUACGGAUAGUUGGGACUGGGCACAUGUAAUGUUGCCGGGCCACCUUGGACGGCUCCUUGCACUCCCACUCCCACCCCAACUC ..(((...)))(((((((((.(((....)))---.)))))).......(((((.((((....(((((.(..((((..((.....)).))))..))))))..)))).))))).)))..... ( -45.70) >DroYak_CAF1 16838 114 + 1 UUGCUUGGUGUGGCGUAUGACCAUAUGGGUGGUGUCCAAAUGGAUAAUUUGGACUCGGAACAUGUAAAGUUGAAGUACUUGUUGGUCCAGUAGUCUGAAC----GCUACCGCU--AAGAC ...(((((((((((((..(((((((((..(.(.((((((((.....)))))))).).)..))))).((((......))))...))))(((....))).))----)))).))))--))).. ( -42.30) >consensus UUGCUUGGGGUGGCGUAUGACCAUAUGGGUGGUGUCCAAAUGGAUAAUUUGGACUCGGAACAUGUAAAGUUGCAGAGCUUGUUGGCCCAGUAGUCUGAAC____GCUACCGCC__AAGAC ......(.((((((........(((((..(.(.((((((((.....)))))))).).)..)))))..((((((.(.(((....))).).)))).))........)))))).)........ (-21.44 = -23.63 + 2.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:42 2006