| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 550,255 – 550,415 |
| Length | 160 |
| Max. P | 0.986628 |

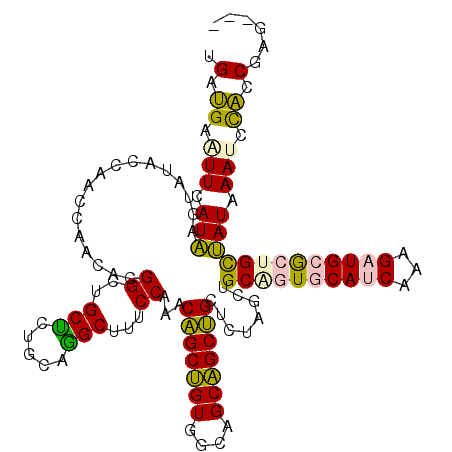
| Location | 550,255 – 550,375 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -24.08 |
| Energy contribution | -22.80 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 550255 120 + 1281640 UGAUGAAUUCAUAACUAUACCAACCAACACGGCUGCCUUGCAGGCUUUCCAAACAGCUGUGGCUGCAGCUGCUCUAGCUGCAGUGCAUCAAAGAUGCGCCGUUAUAAAUCCACCGACACA .(.((.(((.(((((............((((((((((.....))).........)))))))((((((((((...))))))))))(((((...)))))...))))).))).)).)...... ( -36.70) >DroPse_CAF1 3756 117 + 1 UGACGACUUCAUGAGUAUACCAACAAACACGGCGGCUCUGCAAGCCUUCCAAACUGCGGUUGCAGCCGCAGCCAUAGCAGCGGUCCAACAACGCUGUAGUGCCAUCAACAUGCCUAG--- .........((((.((......))...((((((((((.....))))........((((((....)))))))))...((((((.........)))))).))).......)))).....--- ( -33.50) >DroSim_CAF1 1938 120 + 1 UGAUGAGUUUAUAACUAUACCAACCAACACGGCUGCCCUGCAGGCUUUCCAAACAGCUGUGGCUGCAGCUGCUCUAGCUGCCGUGCAUCAAAGAUGCGCUGUUAUAAAUCCACCGAUACA .(.((..((((((((............((((((((((.....))).........)))))))...(((((((...))))))).(((((((...))))))).))))))))..)).)...... ( -37.40) >DroEre_CAF1 3374 116 + 1 UGAUGAAUUUAUAAGUAUACCAACCAAUACGGCUGCUCUGCAGGCUUUCCAAACAGCUGUAGCAGCAGCUGCUCUAGCUGCAGUGCAUCAAAGAUGCGCUGCUAUAAAUCCACCGA---- .(.((.(((((((................((((((((((((((.((........)))))))).))))))))........((((((((((...))))))))))))))))).)).)..---- ( -42.10) >DroPer_CAF1 5378 117 + 1 UGACGACUUCAUGAGUAUACCAACAAACACGGCGGCUCUGCAAGCCUUCCAAACUGCGGUUGCAGCCGCAGCCAUAGCAGCGGUCCAACAACGCUGUAGUGCCAUCAACAUGCCUAG--- .........((((.((......))...((((((((((.....))))........((((((....)))))))))...((((((.........)))))).))).......)))).....--- ( -33.50) >consensus UGAUGAAUUCAUAAGUAUACCAACCAACACGGCUGCUCUGCAGGCUUUCCAAACAGCUGUGGCAGCAGCUGCUCUAGCUGCAGUGCAUCAAAGAUGCGCUGCUAUAAAUCCACCGAG___ .(.((.(((.(((.................((..(((.....)))...))...(((((((....)))))))........((((((((((...))))))))))))).))).)).)...... (-24.08 = -22.80 + -1.28)

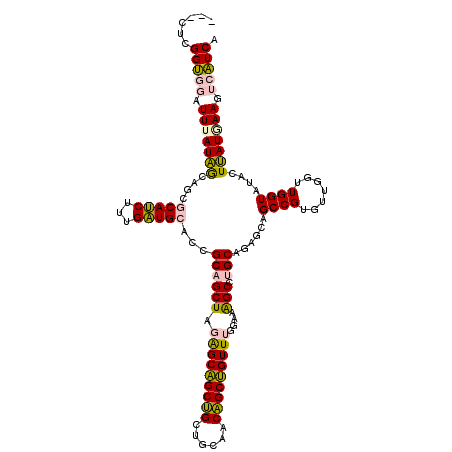
| Location | 550,255 – 550,375 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -43.06 |
| Consensus MFE | -31.12 |
| Energy contribution | -30.32 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 550255 120 - 1281640 UGUGUCGGUGGAUUUAUAACGGCGCAUCUUUGAUGCACUGCAGCUAGAGCAGCUGCAGCCACAGCUGUUUGGAAAGCCUGCAAGGCAGCCGUGUUGGUUGGUAUAGUUAUGAAUUCAUCA ......((((((((((((((((((((((...)))))..(((((((.....)))))))))).((((((..(((...(((.....)))..)))...)))))).....)))))))))))))). ( -51.30) >DroPse_CAF1 3756 117 - 1 ---CUAGGCAUGUUGAUGGCACUACAGCGUUGUUGGACCGCUGCUAUGGCUGCGGCUGCAACCGCAGUUUGGAAGGCUUGCAGAGCCGCCGUGUUUGUUGGUAUACUCAUGAAGUCGUCA ---...(((......(((((....(((((.........)))))(((.((((((((......)))))))))))..(((((...)))))))))).((..(.((....)).)..))...))). ( -35.50) >DroSim_CAF1 1938 120 - 1 UGUAUCGGUGGAUUUAUAACAGCGCAUCUUUGAUGCACGGCAGCUAGAGCAGCUGCAGCCACAGCUGUUUGGAAAGCCUGCAGGGCAGCCGUGUUGGUUGGUAUAGUUAUAAACUCAUCA ......((((..((((((((...(((((...)))))(((((.....(((((((((......))))))))).....(((.....))).))))).............))))))))..)))). ( -45.40) >DroEre_CAF1 3374 116 - 1 ----UCGGUGGAUUUAUAGCAGCGCAUCUUUGAUGCACUGCAGCUAGAGCAGCUGCUGCUACAGCUGUUUGGAAAGCCUGCAGAGCAGCCGUAUUGGUUGGUAUACUUAUAAAUUCAUCA ----..((((((((((((((((.(((((...))))).))))((((((.((.((((((.((.(((((........)).))).)))))))).)).))))))........)))))))))))). ( -47.60) >DroPer_CAF1 5378 117 - 1 ---CUAGGCAUGUUGAUGGCACUACAGCGUUGUUGGACCGCUGCUAUGGCUGCGGCUGCAACCGCAGUUUGGAAGGCUUGCAGAGCCGCCGUGUUUGUUGGUAUACUCAUGAAGUCGUCA ---...(((......(((((....(((((.........)))))(((.((((((((......)))))))))))..(((((...)))))))))).((..(.((....)).)..))...))). ( -35.50) >consensus ___CUCGGUGGAUUUAUAGCAGCGCAUCUUUGAUGCACCGCAGCUAGAGCAGCUGCUGCAACAGCUGUUUGGAAAGCCUGCAGAGCAGCCGUGUUGGUUGGUAUACUUAUGAAGUCAUCA ......(((((.(((((((....(((((...)))))...((((((.(((((((((......)))))))))....))).)))......((((.......))))....))))))).))))). (-31.12 = -30.32 + -0.80)

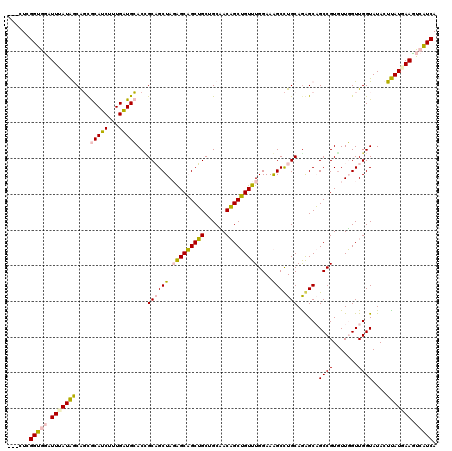
| Location | 550,295 – 550,415 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.05 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.06 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 550295 120 + 1281640 CAGGCUUUCCAAACAGCUGUGGCUGCAGCUGCUCUAGCUGCAGUGCAUCAAAGAUGCGCCGUUAUAAAUCCACCGACACAGGAUACCGUCGAUGAAGACAAGGACUUAGAUACGAAUGUC ..(((................((((((((((...))))))))))(((((...))))))))((..(((.(((.((......)).....(((......)))..))).)))...))....... ( -36.60) >DroPse_CAF1 3796 108 + 1 CAAGCCUUCCAAACUGCGGUUGCAGCCGCAGCCAUAGCAGCGGUCCAACAACGCUGUAGUGCCAUCAACAUGCCUAG------------AGAAGCAGAUACGGAAAUCGGUCCUUGCAUU ((((((((((...(((((((....))))))).(((.((((((.........)))))).))).........(((....------------....))).....))))...))..)))).... ( -30.10) >DroSim_CAF1 1978 120 + 1 CAGGCUUUCCAAACAGCUGUGGCUGCAGCUGCUCUAGCUGCCGUGCAUCAAAGAUGCGCUGUUAUAAAUCCACCGAUACAGGAGACCGUCGACGAAGACAAGGACUUAGAUACGAAUGUC ..(((.(((..((((((....((.(((((((...))))))).))(((((...))))))))))).(((.(((.((......)).....(((......)))..))).))).....))).))) ( -35.80) >DroEre_CAF1 3414 111 + 1 CAGGCUUUCCAAACAGCUGUAGCAGCAGCUGCUCUAGCUGCAGUGCAUCAAAGAUGCGCUGCUAUAAAUCCACCGA---------CCGUCGAGGAAGAUAAGGACAUAGAUACGAAUGUC .....(((((....(((((.(((((...))))).)))))((((((((((...))))))))))..............---------.......))))).....(((((........))))) ( -35.00) >DroPer_CAF1 5418 108 + 1 CAAGCCUUCCAAACUGCGGUUGCAGCCGCAGCCAUAGCAGCGGUCCAACAACGCUGUAGUGCCAUCAACAUGCCUAG------------AGAAGCAGAUACGGAAAUCGGUCCUUGCAUU ((((((((((...(((((((....))))))).(((.((((((.........)))))).))).........(((....------------....))).....))))...))..)))).... ( -30.10) >consensus CAGGCUUUCCAAACAGCUGUGGCAGCAGCUGCUCUAGCUGCAGUGCAUCAAAGAUGCGCUGCUAUAAAUCCACCGAG________CCGUCGAAGAAGAUAAGGACAUAGAUACGAAUGUC .......(((...(((((((....)))))))........((((((((((...)))))))))).......................................)))................ (-21.98 = -22.06 + 0.08)

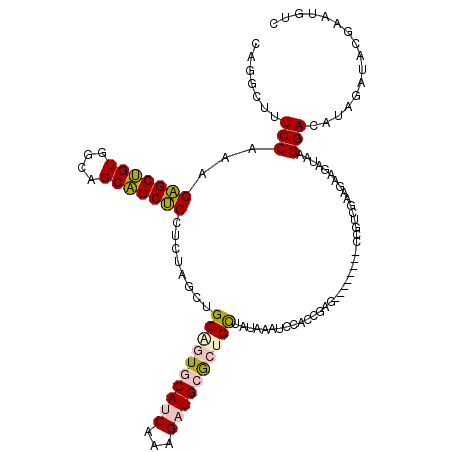
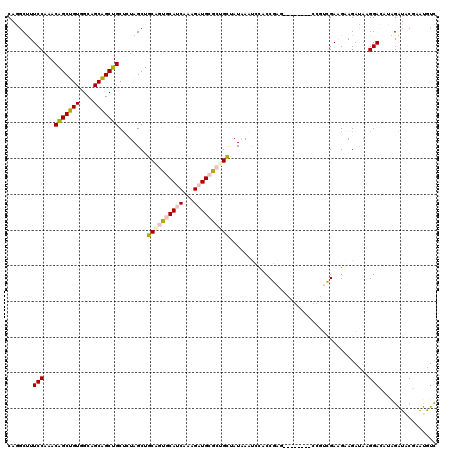
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:35 2006