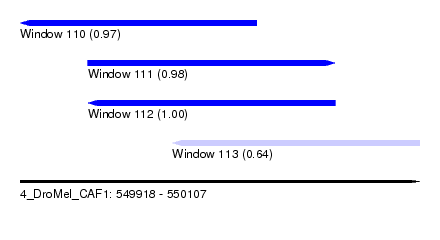
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 549,918 – 550,107 |
| Length | 189 |
| Max. P | 0.995631 |

| Location | 549,918 – 550,030 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.19 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -10.94 |
| Energy contribution | -12.22 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 549918 112 - 1281640 GCAGAUGAUGCCGCAGCAUAGCUGGCAUCCGUUGUGGCAUCAUUGGUUGUGGUUCUAGCCAAAUUUAGGCCAGGUGCGAGAGAAACAUCUG--------AAAACAUAACACCUUAAUAAG ...((((((((((((((.............)))))))))))))).((((((......(((.......)))(((((((....)...))))))--------....))))))........... ( -36.92) >DroPse_CAF1 3410 109 - 1 GC---UGUUGCUGCGGCAAAGCCGACCUCCGACGAUCCACCAACUGGUGUGUUUGUUGCCAUAUUGAAGCCAGCUUCAAAAAUGAUAUCUG--------AAAUAAAGAUCCAAUCGUAAA ((---((..(((.((((...)))).....((((((.(((((....)))).).)))))).........))))))).......(((((((((.--------......))))...)))))... ( -26.40) >DroSim_CAF1 1604 109 - 1 GCAGAUGAUGCCGCAGCAUAGCUGGCAUCCGUU---GCAUCAUUGGUUGCGGUUCUAGCCAAAUUUAGGCCAGAUGCGAAAGAAACAUCUG--------AAAAUAAAACACCUCAAUAAA ((((..((((((((......)).))))))..))---)).......((((.(((....(((.......)))(((((((....)...))))))--------..........))).))))... ( -33.10) >DroEre_CAF1 3026 120 - 1 GCAGAUGAUGCCUCAGCAUAGCUGGCAUCCGACGUGGCAUCAUUGGUUGUGGCUCUAGCCAAAUUAAGGCCAGAUGCAAAAGAAACAUCUGCAAAAAAAAAAAAAAAACACCUUAAUGAG (((((((......((((...))))(((((.....((((....((((((.((((....)))))))))).)))))))))........)))))))............................ ( -35.10) >DroPer_CAF1 5032 109 - 1 GC---UGUUGCUGCGGCAAAGCCGACCUCCGACGAUCCACCAACUGGUGUGUUUGUUGCCAUAUUGAAGCCAGCUUCAAAAAUGAUAUCUG--------AAAUAAAGAUCCAAUCGUAAA ((---((..(((.((((...)))).....((((((.(((((....)))).).)))))).........))))))).......(((((((((.--------......))))...)))))... ( -26.40) >consensus GCAGAUGAUGCCGCAGCAUAGCUGGCAUCCGACGAGGCAUCAUUGGUUGUGGUUCUAGCCAAAUUGAGGCCAGAUGCAAAAGAAACAUCUG________AAAAAAAAACACCUUAAUAAA ...((((((((..((((...))))............))))))))(((..........(((.......)))((((((.........))))))..................)))........ (-10.94 = -12.22 + 1.28)

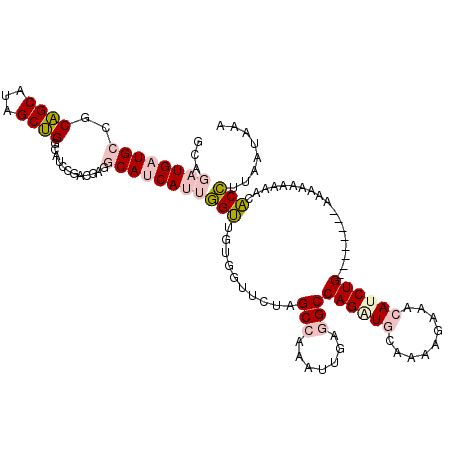
| Location | 549,950 – 550,067 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.39 |
| Mean single sequence MFE | -40.49 |
| Consensus MFE | -18.90 |
| Energy contribution | -17.54 |
| Covariance contribution | -1.36 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 549950 117 + 1281640 CUCGCACCUGGCCUAAAUUUGGCUAGAACCACAACCAAUGAUGCCACAACGGAUGCCAGCUAUGCUGCGGCAUCAUCUGCGGC---AGUUCCUGCCAUUCCGGAUGUCAGUAUGUUUUCA ...(((.(((((.......((((..((((..((.....)).((((.((...((((((.((......))))))))...)).)))---)))))..))))........)))))..)))..... ( -35.96) >DroPse_CAF1 3442 114 + 1 UUUGAAGCUGGCUUCAAUAUGGCAACAAACACACCAGUUGGUGGAUCGUCGGAGGUCGGCUUUGCCGCAGCAACA---GCGGG---AUCCUCCGCCGCUGCAGAUGGCAAUAUAUUAUCU ...((((((((((((....((....))....((((....))))........))))))))))))((((((((....---(((((---....))))).)))))....)))............ ( -43.40) >DroSim_CAF1 1636 114 + 1 UUCGCAUCUGGCCUAAAUUUGGCUAGAACCGCAACCAAUGAUGC---AACGGAUGCCAGCUAUGCUGCGGCAUCAUCUGCGGC---AGCUCCUGCCAUUCCGGAUGUCAGUAUGUUUUCA ...((.(((((((.......)))))))...(((.((......((---(...((((((.((......))))))))...)))(((---((...))))).....)).)))..))......... ( -39.10) >DroEre_CAF1 3066 120 + 1 UUUGCAUCUGGCCUUAAUUUGGCUAGAGCCACAACCAAUGAUGCCACGUCGGAUGCCAGCUAUGCUGAGGCAUCAUCUGCGGCUGGAGCCCCUGCCAUUCCGGAUGUCAAUAUGUUUUCA .((((((((((.........((((..((((.((....((((((((..((.....))((((...)))).)))))))).)).))))..)))).........))))))).))).......... ( -40.57) >DroPer_CAF1 5064 114 + 1 UUUGAAGCUGGCUUCAAUAUGGCAACAAACACACCAGUUGGUGGAUCGUCGGAGGUCGGCUUUGCCGCAGCAACA---GCGGG---AUCCUCCGCCGCUGCAGAUGGCAAUAUAUUAUCU ...((((((((((((....((....))....((((....))))........))))))))))))((((((((....---(((((---....))))).)))))....)))............ ( -43.40) >consensus UUUGCAGCUGGCCUAAAUUUGGCUAGAACCACAACCAAUGAUGCAACGUCGGAUGCCAGCUAUGCUGCGGCAUCAUCUGCGGC___AGCCCCUGCCAUUCCGGAUGUCAAUAUGUUUUCA .......((((((.......)))................((((........(((((((((...))))..)))))....((((.........)))))))).)))................. (-18.90 = -17.54 + -1.36)

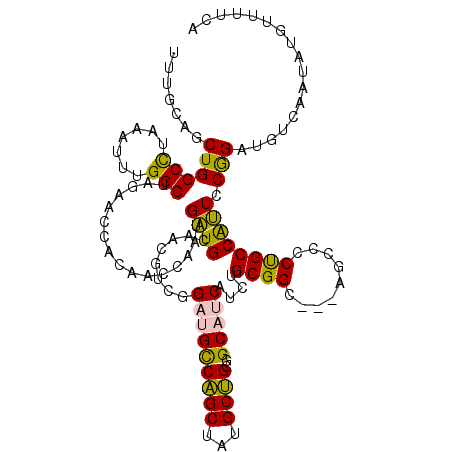
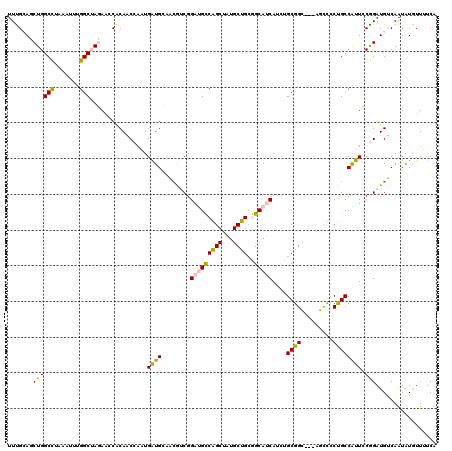
| Location | 549,950 – 550,067 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.39 |
| Mean single sequence MFE | -42.64 |
| Consensus MFE | -21.52 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 549950 117 - 1281640 UGAAAACAUACUGACAUCCGGAAUGGCAGGAACU---GCCGCAGAUGAUGCCGCAGCAUAGCUGGCAUCCGUUGUGGCAUCAUUGGUUGUGGUUCUAGCCAAAUUUAGGCCAGGUGCGAG ...........((.((((.((..((((.(((((.---.(.((.((((((((((((((.............)))))))))))))).)).)..))))).))))........)).)))))).. ( -44.82) >DroPse_CAF1 3442 114 - 1 AGAUAAUAUAUUGCCAUCUGCAGCGGCGGAGGAU---CCCGC---UGUUGCUGCGGCAAAGCCGACCUCCGACGAUCCACCAACUGGUGUGUUUGUUGCCAUAUUGAAGCCAGCUUCAAA ..........(((((....((((((((((.....---.))))---))))))...)))))..........((((((.(((((....)))).).)))))).....((((((....)))))). ( -42.00) >DroSim_CAF1 1636 114 - 1 UGAAAACAUACUGACAUCCGGAAUGGCAGGAGCU---GCCGCAGAUGAUGCCGCAGCAUAGCUGGCAUCCGUU---GCAUCAUUGGUUGCGGUUCUAGCCAAAUUUAGGCCAGAUGCGAA ...........((.((((.((..((((.((((((---((.((.((((((((.(((((...)))((...)))).---)))))))).)).)))))))).))))........)).)))))).. ( -39.20) >DroEre_CAF1 3066 120 - 1 UGAAAACAUAUUGACAUCCGGAAUGGCAGGGGCUCCAGCCGCAGAUGAUGCCUCAGCAUAGCUGGCAUCCGACGUGGCAUCAUUGGUUGUGGCUCUAGCCAAAUUAAGGCCAGAUGCAAA ..........(((.((((.((..((((.(((((..(((((...(((((((((.((((...))))((.......))))))))))))))))..))))).))))........)).))))))). ( -45.20) >DroPer_CAF1 5064 114 - 1 AGAUAAUAUAUUGCCAUCUGCAGCGGCGGAGGAU---CCCGC---UGUUGCUGCGGCAAAGCCGACCUCCGACGAUCCACCAACUGGUGUGUUUGUUGCCAUAUUGAAGCCAGCUUCAAA ..........(((((....((((((((((.....---.))))---))))))...)))))..........((((((.(((((....)))).).)))))).....((((((....)))))). ( -42.00) >consensus UGAAAACAUAUUGACAUCCGGAAUGGCAGGGGCU___GCCGCAGAUGAUGCCGCAGCAUAGCUGGCAUCCGACGAGGCAUCAUUGGUUGUGGUUCUAGCCAAAUUGAGGCCAGAUGCAAA ..........(((....((.(((((((.((((((...((((...(((((((..((((...))))............)))))))))))...)))))).))))..))).)).)))....... (-21.52 = -20.60 + -0.92)


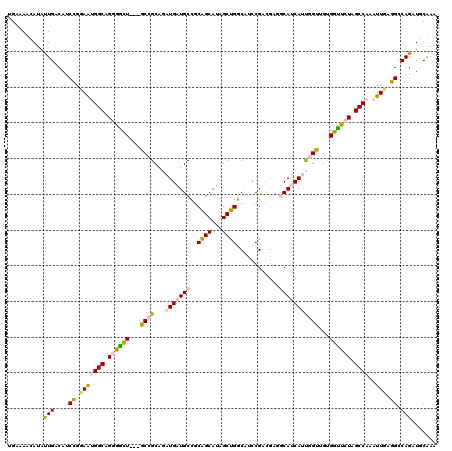
| Location | 549,990 – 550,107 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -41.96 |
| Consensus MFE | -17.26 |
| Energy contribution | -18.86 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 549990 117 - 1281640 UCUUAUCGCAAGAUGUUGCGCAACUUGAGGGAGAUGUUGGUGAAAACAUACUGACAUCCGGAAUGGCAGGAACU---GCCGCAGAUGAUGCCGCAGCAUAGCUGGCAUCCGUUGUGGCAU ...((((....))))((((.((.((....)).(((((..(((......)))..))))).....))))))....(---(((((((..((((((((......)).))))))..)))))))). ( -41.40) >DroPse_CAF1 3482 114 - 1 UCUUGCCACAAGAUGUCGAGCAACUAGAGGUCGACUUUGGAGAUAAUAUAUUGCCAUCUGCAGCGGCGGAGGAU---CCCGC---UGUUGCUGCGGCAAAGCCGACCUCCGACGAUCCAC (((((...)))))(((((........(((((((.((((((..(((...)))..))..((((((((((((.((..---.)).)---))))))))))).)))).))))))))))))...... ( -42.50) >DroSim_CAF1 1676 114 - 1 UCUUAUCGCAAGAUGUUGCGCAACUUGAGGGAGAUGUUGGUGAAAACAUACUGACAUCCGGAAUGGCAGGAGCU---GCCGCAGAUGAUGCCGCAGCAUAGCUGGCAUCCGUU---GCAU ...((((....))))....(((((......(.(((((..(((......)))..))))))(((.((.(((..(((---((.(((.....))).)))))....))).))))))))---)).. ( -44.00) >DroEre_CAF1 3106 120 - 1 UCUUGUCGCAAGAUGUUGCGCAACUUGAGGGAGAUGUUGGUGAAAACAUAUUGACAUCCGGAAUGGCAGGGGCUCCAGCCGCAGAUGAUGCCUCAGCAUAGCUGGCAUCCGACGUGGCAU ((((((((((((.((.....)).))))...(.(((((..(((......)))..))))))....))))))))......(((((....((((((..(((...)))))))))....))))).. ( -39.40) >DroPer_CAF1 5104 114 - 1 UCUUGCCACAAGAUGUCGAGCAACUAGAGGUCGACUUUGGAGAUAAUAUAUUGCCAUCUGCAGCGGCGGAGGAU---CCCGC---UGUUGCUGCGGCAAAGCCGACCUCCGACGAUCCAC (((((...)))))(((((........(((((((.((((((..(((...)))..))..((((((((((((.((..---.)).)---))))))))))).)))).))))))))))))...... ( -42.50) >consensus UCUUGUCGCAAGAUGUUGCGCAACUUGAGGGAGAUGUUGGUGAAAACAUAUUGACAUCCGGAAUGGCAGGGGCU___GCCGCAGAUGAUGCCGCAGCAUAGCUGGCAUCCGACGAGGCAU (((((((.((((.((.....)).)))).....((((((((((......))))))))))......))))))).........((...(((((...((((...)))).....)))))..)).. (-17.26 = -18.86 + 1.60)

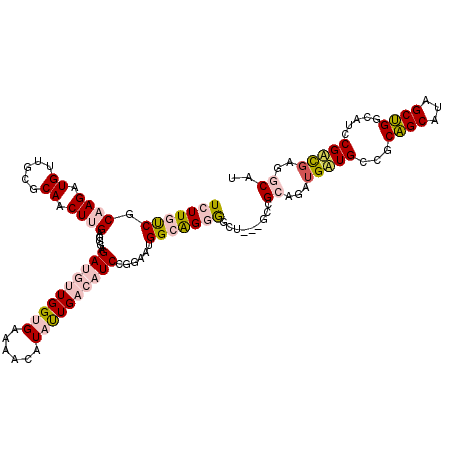
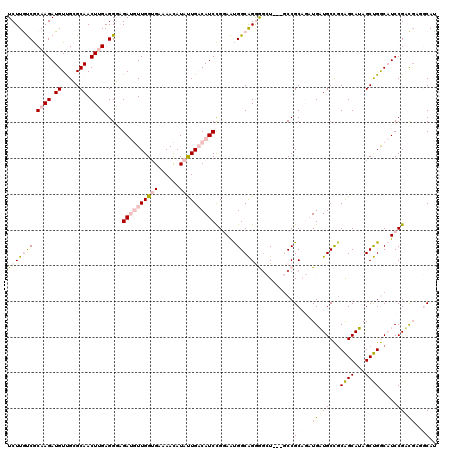
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:31 2006