| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 538,223 – 538,324 |
| Length | 101 |
| Max. P | 0.985393 |

| Location | 538,223 – 538,324 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.65 |
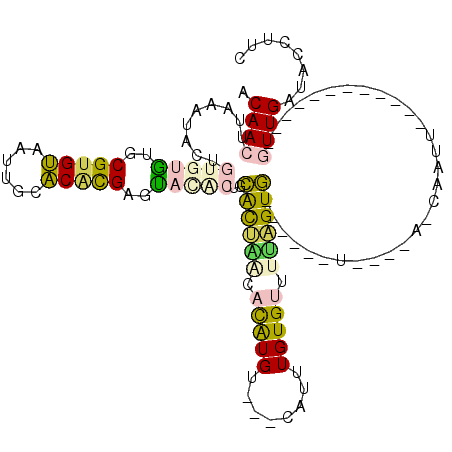
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -15.02 |
| Energy contribution | -14.33 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.64 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 538223 101 + 1281640 ACAACUUAAAUACUGUGUGUGCGUGUAAUUGCACACGAGUACACUGCACUAGCACAUGU---CAUUUGUGUUUUAGUG------U----A-CAAUAGUG-----UACAGUUGAUACCUUC .(((((...((((((((((((((......)))))))..((((((((....((((((...---....)))))).)))))------)----)-).))))))-----)..)))))........ ( -36.70) >DroPse_CAF1 35227 100 + 1 ACAACUUAAAUAUUAU-UAUGCGUGCUGUUUCGCAUGUGUUU-CUGUGUGGUUCUGUGUAAUUAUGUGUGUAUUCGUA------UGUGUAUCAGCU------------AUUGUUGCAUUC ..((((...((((...-.((((((((......))))))))..-..))))))))..(((((((...((..((((.....------...))))..)).------------...))))))).. ( -16.80) >DroSec_CAF1 10813 94 + 1 ACAACUUAAAUACUGUGUGUGCGAGUAAUUGCACCCGAGUACGCUGCACUAGCACAUGU---CAUUUGUGUUUUAGUG------U----A-CAGUU------------GUUGAUACCUUC ((((((.........((.((((((....)))))).)).((((((((....((((((...---....)))))).)))))------)----)-)))))------------)).......... ( -28.50) >DroEre_CAF1 37436 112 + 1 ACAACUUAAAUACUGUGUGUGCGUGUAAUUGCACACGAGCAUACUGCACUAACACAUGC---CAUUUGUGUUUAUGUGGGCGAUU----G-CGAUUGCUACCAGAAUUGUUGAUACCGUC .((((...(((.(((((((((((......))))))))((((..(..((..((((((...---....))))))..))..)((....----)-)...))))..))).)))))))........ ( -31.30) >DroYak_CAF1 10389 106 + 1 ACAACUUAAAUACUGUGUGUGCGUGUAAUUGCACGUGAGCACACUGCACUCACACAUGU---CAUUUGUGUUUGAGUG------U----A-CGAUUGCUACCACAAUUGUUGAUACCUUC .((((.........((((((((((((....))))))..))))))((((((((.(((((.---....))))).))))))------)----)-.(((((......)))))))))........ ( -33.40) >DroPer_CAF1 35336 100 + 1 ACAACUUAAAUAUUAU-UAUGCGUGCUGUUUCGCAUGUGUUU-CUGUGUGGUUCUGUGUAAUUAUGUGUGUAUUCGUA------UGUGUAUCAGCU------------AUUGUUGCAUUC ..((((...((((...-.((((((((......))))))))..-..))))))))..(((((((...((..((((.....------...))))..)).------------...))))))).. ( -16.80) >consensus ACAACUUAAAUACUGUGUGUGCGUGUAAUUGCACACGAGUACACUGCACUAACACAUGU___CAUUUGUGUUUUAGUG______U____A_CAAUU____________GUUGAUACCUUC .((((.........(((((..(((((......)))))..)))))..((((((.(((((........))))).))))))..............................))))........ (-15.02 = -14.33 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:25 2006