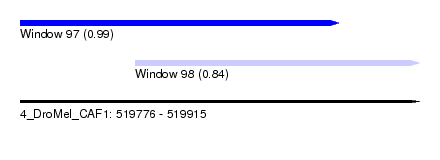
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 519,776 – 519,915 |
| Length | 139 |
| Max. P | 0.994816 |

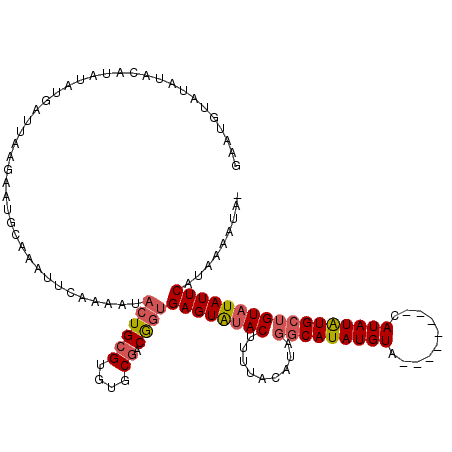
| Location | 519,776 – 519,887 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -13.26 |
| Energy contribution | -14.20 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 519776 111 + 1281640 GUACGUACAUACGUAUAUGAUUAAAAAUUCAAAUUCAAAAUACUGCGUGUGCGACGGUGAGUACACUUUUACAUAGGCAUAUGUA--------CAUAUAUGCUGUAUAUUCAUAAAAUA- ((.((((((..(((((.(((..............)))..)))).)..)))))))).(((((((.((.........((((((((..--------..)))))))))).)))))))......- ( -23.04) >DroSec_CAF1 26153 111 + 1 GAAUGUAUAUACAUAUAUGAUUAAGAAUGCUAAUUCAAAGUACUGCGUGUGCGACGGUGAGUAUACUUUUACAUAGGCAUAUGUA--------CAUAUAUGCUGUAUAUUCAUAAAAUA- (((((((((..((((((((.....((((....)))).......(((((((((....(((((......)))))....)))))))))--------)))))))).)))))))))........- ( -32.00) >DroSim_CAF1 26968 111 + 1 GAAUGUAUAUACAUAUAUGAUUAAGAAUGCUAAUUCAAAGUACUGCGUGUGCGACGGUGAGUAUACUUUUACAUAGGCAUAUGUA--------CAUAUAUGCUGUAUAUUCAUAAAAUA- (((((((((..((((((((.....((((....)))).......(((((((((....(((((......)))))....)))))))))--------)))))))).)))))))))........- ( -32.00) >DroYak_CAF1 32598 99 + 1 ---------------------GUAUAUUGCAAAUGCAAAAUCCUGAGCGUGCGACAUUGAGUGUACUUUUACAUGUACAUAUGUAUGUAUCCACAUAUGUGCUGUAUAUUCAUACUAUCU ---------------------((((.(((((....((......))....)))))....((((((((........((((((((((........)))))))))).))))))))))))..... ( -24.90) >consensus GAAUGUAUAUACAUAUAUGAUUAAGAAUGCAAAUUCAAAAUACUGCGUGUGCGACGGUGAGUAUACUUUUACAUAGGCAUAUGUA________CAUAUAUGCUGUAUAUUCAUAAAAUA_ .........................................((((((....)).))))((((((((.........(((((((((..........)))))))))))))))))......... (-13.26 = -14.20 + 0.94)

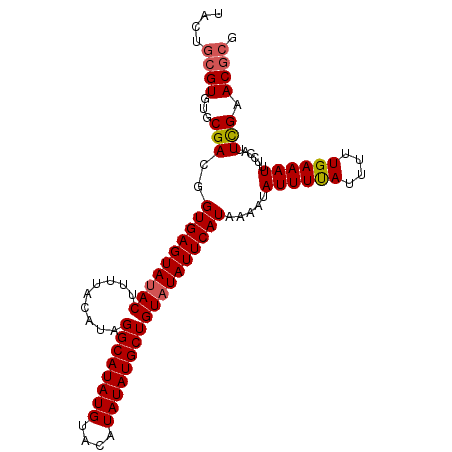
| Location | 519,816 – 519,915 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -21.31 |
| Energy contribution | -21.87 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 519816 99 + 1281640 UACUGCGUGUGCGACGGUGAGUACACUUUUACAUAGGCAUAUGUACAUAUAUGCUGUAUAUUCAUAAAAUAUUUUAUUUUUGAAAUUACCAUUUGAACU-- ...(((((((((........)))))).........((((((((....)))))))))))..((((......((((((....)))))).......))))..-- ( -19.32) >DroSec_CAF1 26193 101 + 1 UACUGCGUGUGCGACGGUGAGUAUACUUUUACAUAGGCAUAUGUACAUAUAUGCUGUAUAUUCAUAAAAUAUUUUAUUUUUGAAAUUUCCAUUCGAACGCG ....((((...(((..((((((((((.........((((((((....)))))))))))))))))).....((((((....))))))......))).)))). ( -25.30) >DroSim_CAF1 27008 101 + 1 UACUGCGUGUGCGACGGUGAGUAUACUUUUACAUAGGCAUAUGUACAUAUAUGCUGUAUAUUCAUAAAAUAUUUCAUUUUUGAAAUUUCCAUUCGAACGCG ....((((...(((..((((((((((.........((((((((....)))))))))))))))))).....((((((....))))))......))).)))). ( -27.50) >consensus UACUGCGUGUGCGACGGUGAGUAUACUUUUACAUAGGCAUAUGUACAUAUAUGCUGUAUAUUCAUAAAAUAUUUUAUUUUUGAAAUUUCCAUUCGAACGCG ....((((...(((..((((((((((.........((((((((....)))))))))))))))))).....((((((....))))))......))).)))). (-21.31 = -21.87 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:09 2006