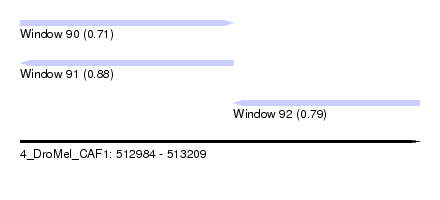
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 512,984 – 513,209 |
| Length | 225 |
| Max. P | 0.879486 |

| Location | 512,984 – 513,104 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -30.91 |
| Consensus MFE | -26.59 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 512984 120 + 1281640 UUUACUGGAGAGCGUAAUUACGAGGAGUACUCUACGCGUUCUGCGACACUUACUGCUCCCAAGCAGCCAAUUCGAAAUACAUGCAUCAACAUAUUAUUUGUGCUGGCUGCUAAUUCUCUU .....(((.((((((((....(((.....)))..(((.....)))....)))).)))))))((((((((...((((....(((......)))....))))...))))))))......... ( -33.30) >DroSec_CAF1 19677 120 + 1 UUAACUGGAGAGCGUAAUUACGAGGAGUACUCUACGCGUUCUGCGACACUUACUACUCCCAAGCAGCCAAUUCGAAAUACAUGCAUCAACAUAUUAUUUGUGCUGGCUGCUAAUUCUCUU ......((((((.........(.((((((.....(((.....)))........))))))).((((((((...((((....(((......)))....))))...))))))))..)))))). ( -30.02) >DroSim_CAF1 19550 120 + 1 UUAACUGGAGAGCGUAAUUACGGGGAGUACUCUACGCGUUCUGCGACACUUACUACUCCCAAGCAGCCAAUUCGAAAUACAUGCAUCAACAUAUUAUUUGUGCUGGCUGCUAAUUCUCUU ......(((((((((....)))(((((((.....(((.....)))........))))))).((((((((...((((....(((......)))....))))...))))))))..)))))). ( -34.12) >DroYak_CAF1 25071 120 + 1 UUUACUGGAGAGCCUAAUUACGAGAAGUACUCUAUGCGUUCUGCGACACGUACUGUUCCCAAGCAGCGAACUCGAAAUACAUGCAUCGACAUAUUAUUUCUGCUGGCUGCUGAUUCUCUU ......((((((.........(((.(((((....(((.....)))....))))).)))...((((((.(.(..((((((.(((......)))..)))))).).).))))))..)))))). ( -26.20) >consensus UUAACUGGAGAGCGUAAUUACGAGGAGUACUCUACGCGUUCUGCGACACUUACUACUCCCAAGCAGCCAAUUCGAAAUACAUGCAUCAACAUAUUAUUUGUGCUGGCUGCUAAUUCUCUU ......((((((.........(((.((((.....(((.....))).....)))).)))...((((((((...((((....(((......)))....))))...))))))))..)))))). (-26.59 = -26.52 + -0.06)

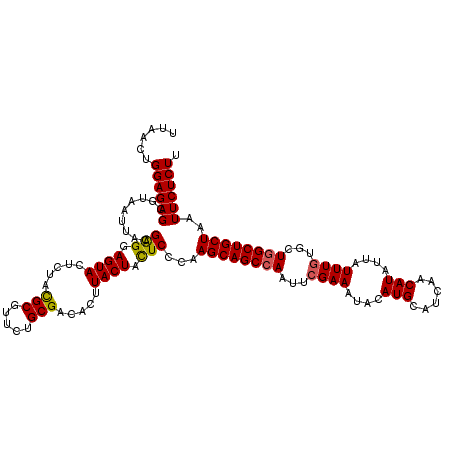
| Location | 512,984 – 513,104 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -30.33 |
| Energy contribution | -30.95 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 512984 120 - 1281640 AAGAGAAUUAGCAGCCAGCACAAAUAAUAUGUUGAUGCAUGUAUUUCGAAUUGGCUGCUUGGGAGCAGUAAGUGUCGCAGAACGCGUAGAGUACUCCUCGUAAUUACGCUCUCCAGUAAA .........(((((((((..((((((.(((((....)))))))))).)..)))))))))(((((((.((((((((......))))...(((.....)))....)))))))).)))..... ( -35.90) >DroSec_CAF1 19677 120 - 1 AAGAGAAUUAGCAGCCAGCACAAAUAAUAUGUUGAUGCAUGUAUUUCGAAUUGGCUGCUUGGGAGUAGUAAGUGUCGCAGAACGCGUAGAGUACUCCUCGUAAUUACGCUCUCCAGUUAA ..((((...(((((((((..((((((.(((((....)))))))))).)..))))))))).(((((((.(..((((......))))....).)))))))...........))))....... ( -35.70) >DroSim_CAF1 19550 120 - 1 AAGAGAAUUAGCAGCCAGCACAAAUAAUAUGUUGAUGCAUGUAUUUCGAAUUGGCUGCUUGGGAGUAGUAAGUGUCGCAGAACGCGUAGAGUACUCCCCGUAAUUACGCUCUCCAGUUAA ..((((...(((((((((..((((((.(((((....)))))))))).)..))))))))).(((((((.(..((((......))))....).)))))))...........))))....... ( -37.90) >DroYak_CAF1 25071 120 - 1 AAGAGAAUCAGCAGCCAGCAGAAAUAAUAUGUCGAUGCAUGUAUUUCGAGUUCGCUGCUUGGGAACAGUACGUGUCGCAGAACGCAUAGAGUACUUCUCGUAAUUAGGCUCUCCAGUAAA ..((((..(((((((.(((.((((((.(((((....)))))))))))..))).)))))).((((..(((((.(((.((.....)))))..))))))))).......)..))))....... ( -35.40) >consensus AAGAGAAUUAGCAGCCAGCACAAAUAAUAUGUUGAUGCAUGUAUUUCGAAUUGGCUGCUUGGGAGCAGUAAGUGUCGCAGAACGCGUAGAGUACUCCUCGUAAUUACGCUCUCCAGUAAA ..((((...(((((((((...(((((.(((((....))))))))))....))))))))).(((((((.(..((((......))))....).)))))))...........))))....... (-30.33 = -30.95 + 0.62)

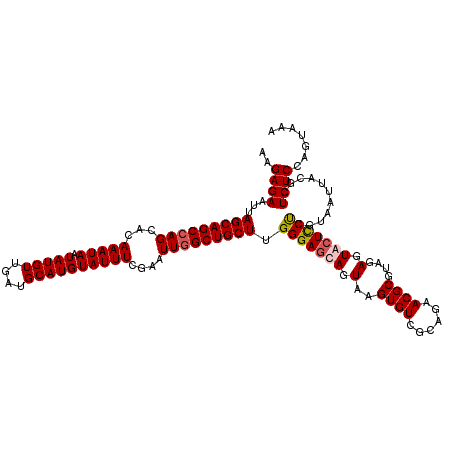
| Location | 513,104 – 513,209 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -19.54 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
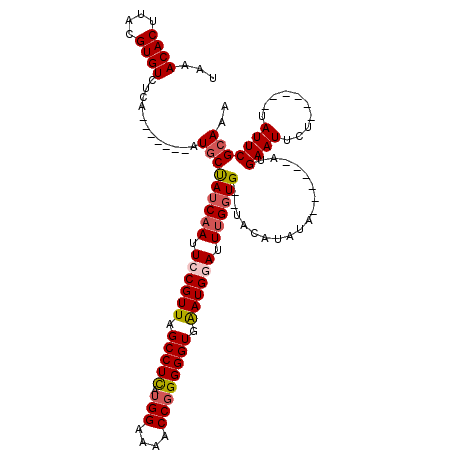
>4_DroMel_CAF1 513104 105 - 1281640 CAAACACUUACGUGUCUCA-------AUUCCAUCAAUUCCGUUAGCCUCAUGGAAAACCGGGGGUGAAUGGAUUUGGUG--UACAUAUA------AUGAAUUCUUAUUCUUAUUCGCAAA ...((((....))))....-------....((((((.((((((.(((((.(((....)))))))).)))))).))))))--.....(((------(.((((....))))))))....... ( -26.50) >DroSec_CAF1 19797 99 - 1 UAAACACUUACGUGUCUCA-------AUGCUAUCAAUUGCGUUAGCCUCAUGGAAAACCGGGGGUGAAUGGAUUUGGUG--UACAUAUA------AUGAAUUCU------UAUUCGCAAA ...((((....))))....-------.(((((((((.(.((((.(((((.(((....)))))))).)))).).))))))--........------..((((...------.))))))).. ( -23.30) >DroSim_CAF1 19670 99 - 1 UAAACACUUACGUGUCUCA-------AUGCUAUCAAUUGCGUUAGCCUCAUGGAAAACCGGGGGUGAAUGGAUUUGGUG--UACAUAUA------AUGAAUUCU------UAUUCGCAAA ...((((....))))....-------.(((((((((.(.((((.(((((.(((....)))))))).)))).).))))))--........------..((((...------.))))))).. ( -23.30) >DroYak_CAF1 25191 113 - 1 UAAACACUUUUGUGUCUUAAUAAUUAAUGCUAUCAAUUCCGUUAGCCUUAGGGAAAACCGGGGGUGGAUGGAUUCG-UGAAUAGAUUUCGUGAAUAUGAAUUCU------UAUUCGCAAA ...((((....))))......................((((((.((((..((.....))..)))).))))))...(-((((((((.(((((....))))).)).------)))))))... ( -31.00) >consensus UAAACACUUACGUGUCUCA_______AUGCUAUCAAUUCCGUUAGCCUCAUGGAAAACCGGGGGUGAAUGGAUUUGGUG__UACAUAUA______AUGAAUUCU______UAUUCGCAAA ...((((....))))............(((((((((.((((((.(((((.(((....)))))))).)))))).))))))..................((((..........))))))).. (-19.54 = -20.47 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:57 2006