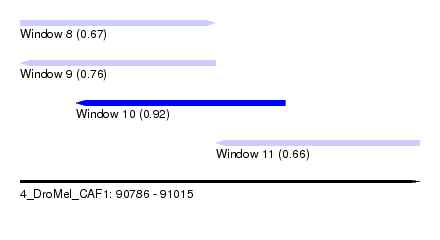
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 90,786 – 91,015 |
| Length | 229 |
| Max. P | 0.924328 |

| Location | 90,786 – 90,898 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.51 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -19.31 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 90786 112 + 1281640 GAUACUCAUCAGAUAGAU----AUAUGUAUA----CGUGUAACUAUGCAUGUAUAUGUAAACACGGCAUUGGAUCCUCUGCUUCGAAUGCAGAGAGCCGACAGUACUCACACACACCCUG ((((((............----.((((((((----((((((....))))))))))))))....((((........((((((.......)))))).))))..)))).))............ ( -29.20) >DroSec_CAF1 36515 112 + 1 GAUACUCAUAAGAUAGAU----AUAUGUAUA----CGUAUAACUAUGCAUGUAUAUGUAAACACGGGAUCGGAUCCUAUGCUUCGAAUGCAGAGAGCCGACAGUACUCACACACACCCUU .................(----(((((((((----.((((....)))).)))))))))).....(((.((((.((...(((.......)))..)).))))..((......))...))).. ( -23.40) >DroEre_CAF1 51151 120 + 1 GAUACCCAUGAGCUAUACAUGCAUAUGUACACGUACGUACUCCUAUGCAUGUAUAUGUAAACACGGGAUCGGAUCCUCUGCUUCGAAUGCAGAGAGCCGACAUUACUCACACACACCCUG ....(((.((.((((((((((((((.((((......))))...)))))))))))).))...)).))).((((...((((((.......))))))..)))).................... ( -40.20) >DroYak_CAF1 58938 114 + 1 GACAUUCAUGAGAUAUA------UAUGUACACACGCGUACACCGCUGCAUGUAUAUGUAAACACGGGAUCGGAUCCUCUGCUUCGAGUGCAGAGAGCCGACAGUACUCACACACACCCUG ........((((...((------((((((((((.(((.....)))))..))))))))))...((..(.((((...((((((.......))))))..))))).)).))))........... ( -31.40) >consensus GAUACUCAUGAGAUAGAU____AUAUGUACA____CGUACAACUAUGCAUGUAUAUGUAAACACGGGAUCGGAUCCUCUGCUUCGAAUGCAGAGAGCCGACAGUACUCACACACACCCUG ......................(((((((((....((((....))))..)))))))))......(((.((((...((((((.......))))))..))))..((......))...))).. (-19.31 = -20.25 + 0.94)

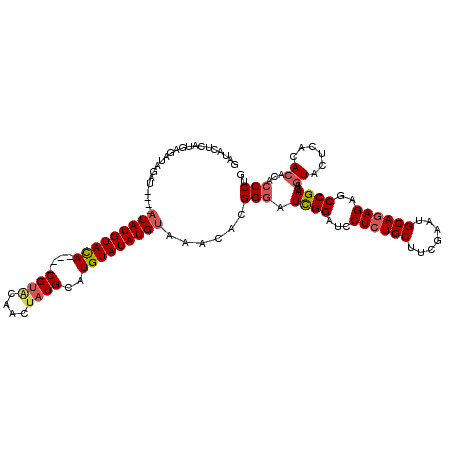
| Location | 90,786 – 90,898 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.51 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -26.25 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 90786 112 - 1281640 CAGGGUGUGUGUGAGUACUGUCGGCUCUCUGCAUUCGAAGCAGAGGAUCCAAUGCCGUGUUUACAUAUACAUGCAUAGUUACACG----UAUACAUAU----AUCUAUCUGAUGAGUAUC ((((..(((((((.(((((((.((.(((((((.......)))))))..)).((((.((((........))))))))....))).)----))).)))))----))...))))......... ( -31.20) >DroSec_CAF1 36515 112 - 1 AAGGGUGUGUGUGAGUACUGUCGGCUCUCUGCAUUCGAAGCAUAGGAUCCGAUCCCGUGUUUACAUAUACAUGCAUAGUUAUACG----UAUACAUAU----AUCUAUCUUAUGAGUAUC ....((((((((((((((.(((((.(((.(((.......)))..))).)))))...))))))))))))))((((.(((.((((.(----....).)))----).)))((....)))))). ( -27.30) >DroEre_CAF1 51151 120 - 1 CAGGGUGUGUGUGAGUAAUGUCGGCUCUCUGCAUUCGAAGCAGAGGAUCCGAUCCCGUGUUUACAUAUACAUGCAUAGGAGUACGUACGUGUACAUAUGCAUGUAUAGCUCAUGGGUAUC ...((((..(((((((...(((((.(((((((.......)))))))..)))))............((((((((((((...(((((....))))).)))))))))))))))))))..)))) ( -44.10) >DroYak_CAF1 58938 114 - 1 CAGGGUGUGUGUGAGUACUGUCGGCUCUCUGCACUCGAAGCAGAGGAUCCGAUCCCGUGUUUACAUAUACAUGCAGCGGUGUACGCGUGUGUACAUA------UAUAUCUCAUGAAUGUC ..(((((((((((..(((.(((((.(((((((.......)))))))..)))))...)))........(((((((.(((.....))))))))))))))------))))))).......... ( -37.70) >consensus CAGGGUGUGUGUGAGUACUGUCGGCUCUCUGCAUUCGAAGCAGAGGAUCCGAUCCCGUGUUUACAUAUACAUGCAUAGGUAUACG____UAUACAUAU____AUAUAUCUCAUGAGUAUC ....((((((((((((((.(((((.(((((((.......)))))))..)))))...)))))))))))))).................................................. (-26.25 = -27.00 + 0.75)

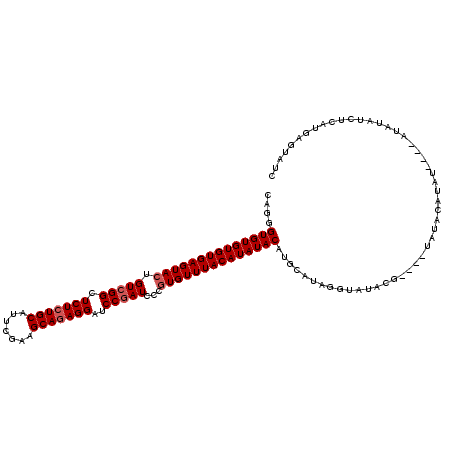
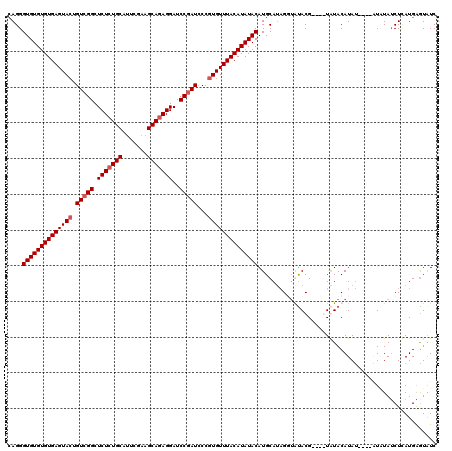
| Location | 90,818 – 90,938 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -30.65 |
| Energy contribution | -31.40 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 90818 120 - 1281640 UACAGAAAACUGUAUUUUCAGCAAAUAAAUAAACAAACAUCAGGGUGUGUGUGAGUACUGUCGGCUCUCUGCAUUCGAAGCAGAGGAUCCAAUGCCGUGUUUACAUAUACAUGCAUAGUU (((((....)))))............................(.(((((((((...((...(((((((((((.......))))))).......)))).))...))))))))).)...... ( -30.91) >DroSec_CAF1 36547 120 - 1 UACAGAAAACUGUAUUUUCAGCAAAUAAAUAAACAAACAUAAGGGUGUGUGUGAGUACUGUCGGCUCUCUGCAUUCGAAGCAUAGGAUCCGAUCCCGUGUUUACAUAUACAUGCAUAGUU (((((....)))))......(((..............(....).((((((((((((((.(((((.(((.(((.......)))..))).)))))...)))))))))))))).)))...... ( -28.80) >DroEre_CAF1 51191 120 - 1 UACAGAAAACUGUAUUUUCAGCAAAUAAAUAAACAAACAUCAGGGUGUGUGUGAGUAAUGUCGGCUCUCUGCAUUCGAAGCAGAGGAUCCGAUCCCGUGUUUACAUAUACAUGCAUAGGA (((((....)))))......(((..............(....).(((((((((((((..(((((.(((((((.......)))))))..)))))....))))))))))))).)))...... ( -32.40) >DroYak_CAF1 58972 120 - 1 UACAGAAAACUGUAUUUUCAGCAAAUAAACAAACAAACAUCAGGGUGUGUGUGAGUACUGUCGGCUCUCUGCACUCGAAGCAGAGGAUCCGAUCCCGUGUUUACAUAUACAUGCAGCGGU (((((....)))))......(((..............(....).((((((((((((((.(((((.(((((((.......)))))))..)))))...)))))))))))))).)))...... ( -34.50) >consensus UACAGAAAACUGUAUUUUCAGCAAAUAAAUAAACAAACAUCAGGGUGUGUGUGAGUACUGUCGGCUCUCUGCAUUCGAAGCAGAGGAUCCGAUCCCGUGUUUACAUAUACAUGCAUAGGU (((((....)))))......(((..............(....).((((((((((((((.(((((.(((((((.......)))))))..)))))...)))))))))))))).)))...... (-30.65 = -31.40 + 0.75)

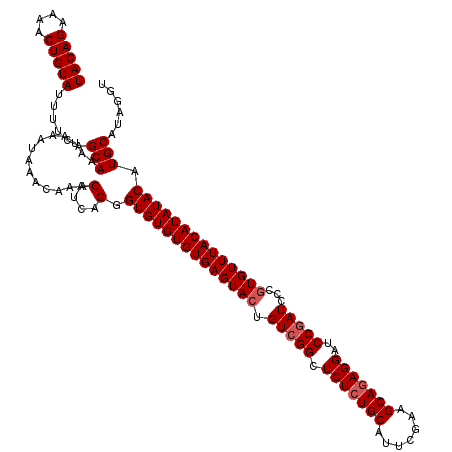
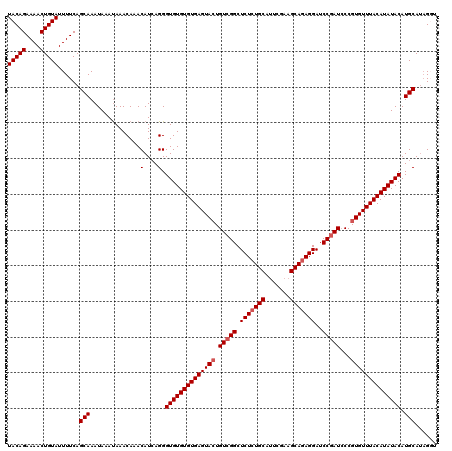
| Location | 90,898 – 91,015 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -17.99 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.97 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 90898 117 - 1281640 GAAACAAGAUUGUGGUGUAUCAACAGGUAGAGACGAAAACGACAGAGUCUAAACAA---AAAUAUAUGCGUAAAACAGAAUACAGAAAACUGUAUUUUCAGCAAAUAAAUAAACAAACAU .........((((.((((((......((..((((............))))..))..---....)))))).......(((((((((....)))))))))..))))................ ( -17.10) >DroSec_CAF1 36627 117 - 1 AAAACAAGAUUGUGGUGUAUCAACAGGUAGAGACGAAAACGACAGAGUCUAAACAA---AAAUAUAUGCGUAAAACAGAAUACAGAAAACUGUAUUUUCAGCAAAUAAAUAAACAAACAU .........((((.((((((......((..((((............))))..))..---....)))))).......(((((((((....)))))))))..))))................ ( -17.10) >DroEre_CAF1 51271 116 - 1 GAGACAAGAUUAUGGCGUAUCAACAGGUAGAGACGGAAACGACA--GCCUAAACAAA--AAAUAUAUGCGUAAAACAGAAUACAGAAAACUGUAUUUUCAGCAAAUAAAUAAACAAACAU .......(.(((.(((.((((....))))....((....))...--)))))).)...--...(((.(((.......(((((((((....)))))))))..))).)))............. ( -19.10) >DroYak_CAF1 59052 118 - 1 GAGACGAGAUUGUGGCGUAUCAACAGGUAGAGACGGAAACGAAA--GUCUAAACAAAAAAAAUAUAUGCGUAAAACAGAAUACAGAAAACUGUAUUUUCAGCAAAUAAACAAACAAACAU .........((((.((((((......((..(((((....)....--))))..)).........)))))).......(((((((((....)))))))))..))))................ ( -18.66) >consensus GAAACAAGAUUGUGGCGUAUCAACAGGUAGAGACGAAAACGACA__GUCUAAACAA___AAAUAUAUGCGUAAAACAGAAUACAGAAAACUGUAUUUUCAGCAAAUAAAUAAACAAACAU ......(((((.((...((((....))))....((....)).)).)))))............(((.(((.......(((((((((....)))))))))..))).)))............. (-13.98 = -14.97 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:56 2006