| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 504,043 – 504,145 |
| Length | 102 |
| Max. P | 0.626356 |

| Location | 504,043 – 504,145 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 89.68 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -21.21 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
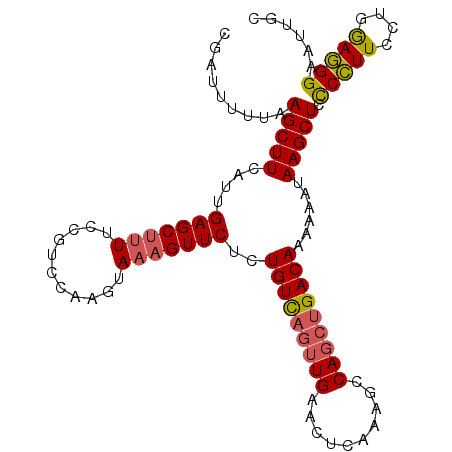
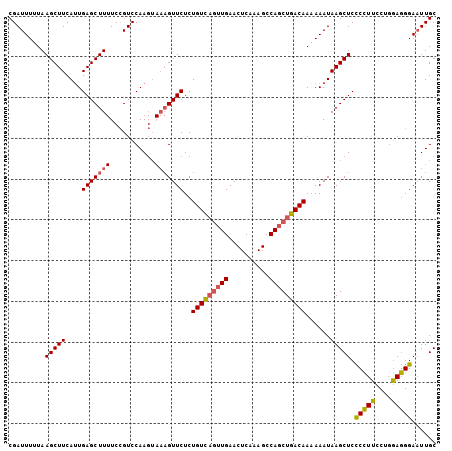
>4_DroMel_CAF1 504043 102 + 1281640 CGAUUUUUAAGCUUCAUUGAGCUUUUCCGUCCAAGUAAAGUUCUCUGUC---UGAACUCAAAGCCAGCUGACAACAAAUAAGCUCCCCUUCCUGGAGGGAAUUGC ........((((((....))))))((((.((((.....(((((......---.))))).......((((...........))))........)))).)))).... ( -21.40) >DroSec_CAF1 10334 105 + 1 CGAUUUUUAAGCUUCAUUGAGCUUUUCCGUCCAAGUAAAGUUCUCUGUUAGUUGAACUCAAAGCCAGCUGACAAAAAAUAAGCUCCCCUUCCUGGAGGGAAUUGC (((((....(((((.((((((((((..(......).)))))))..(((((((((..........)))))))))...)))))))).(((((....)))))))))). ( -26.00) >DroSim_CAF1 10387 105 + 1 CGAUUUUUAAGCUUCAUUGAGCUUUUCCGUCCAAGUAAAGUUCUCUGUUAGUUGAACUCAAAGCCAGCUGACAAAAAAUAAGCUCCCCUUCCUGGAGGGACUUGC .(((....((((((....))))))....)))(((((.........(((((((((..........)))))))))............(((((....)))))))))). ( -26.90) >DroYak_CAF1 14703 99 + 1 CGAUUUGUAAGCUUCAGUGAGCUU-UCUGUCCAA-----GUUCUCUGUCAGUUGAACUCAAAGCCAGCUGACAACAAAUAAGCUCUCUUUCCAAAAAGGAAUUGC ......(..(((((.((.((((((-.......))-----)))).))((((((((..........)))))))).......)))))..).((((.....)))).... ( -23.30) >consensus CGAUUUUUAAGCUUCAUUGAGCUUUUCCGUCCAAGUAAAGUUCUCUGUCAGUUGAACUCAAAGCCAGCUGACAAAAAAUAAGCUCCCCUUCCUGGAGGGAAUUGC .........(((((....(((((((...........)))))))..(((((((((..........)))))))))......))))).(((((....)))))...... (-21.21 = -21.52 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:47 2006