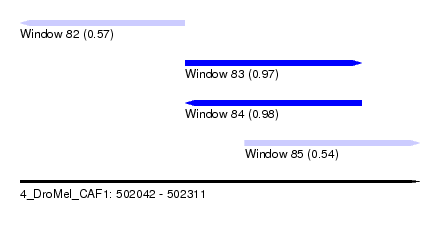
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 502,042 – 502,311 |
| Length | 269 |
| Max. P | 0.980909 |

| Location | 502,042 – 502,153 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.46 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -14.64 |
| Energy contribution | -16.63 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 502042 111 - 1281640 UCUUUUCCUUCCCUAGUGCGU-AUAGAGCGCUCAAAAUGAUCUAUAUUUUUCGUAUUAUAGACAACUGAUGUUUUG--------GCAUUUACUAUGCGUUCCGUAAGUCGCUCUGAAGAU .......((((...(((((.(-(..((((((......((.((((((..........))))))))...(((((....--------)))))......))))))..)).).))))..)))).. ( -23.70) >DroSec_CAF1 8324 102 - 1 -----------------GCGUUUUAGAGCGCCCAAAAUGAUCUGUAUAUUCCGU-UUAUAGUCAACUGAUGAUUUGGGCUAUAGGCAUUUACUAUGCGUUCCGUAAGUCGCUCUAAAGAU -----------------...((((((((((((((((.(((.(((((........-.)))))))).(....).))))))).....((((.....))))............))))))))).. ( -28.50) >DroSim_CAF1 8479 102 - 1 -----------------GCGUUUUAGAGCGCCCAAAAUGAUCUGUAUAUUCCGU-UUAUAGUCAACUGAUGAUUUGGGCUAUAGGCAUUUACUAUGCGUUCCGUAAGUCGUUCUAAAGAU -----------------...((((((((((((((((.(((.(((((........-.)))))))).(....).))))))).....((((.....))))............))))))))).. ( -26.10) >DroYak_CAF1 12864 101 - 1 UUUAUUCCCUCCCUGGUGCGUUAUAGAGCGCCCUAAAUGAUCUAUAU---------CAUAUUUAACUGAUGUUUUGGG----------UUAAUAUGGUGCCCGUAAGUUGCUCUAAGGAU .........(((.(((.((((((..(.(((((.....((((((((((---------((........))))))...)))----------)))....))))))..)))..))).))).))). ( -24.20) >consensus _________________GCGUUAUAGAGCGCCCAAAAUGAUCUAUAUAUUCCGU_UUAUAGUCAACUGAUGAUUUGGG______GCAUUUACUAUGCGUUCCGUAAGUCGCUCUAAAGAU ....................((((((((((((((((((.(((.........................)))))))))))).............((((.....))))....))))))))).. (-14.64 = -16.63 + 2.00)

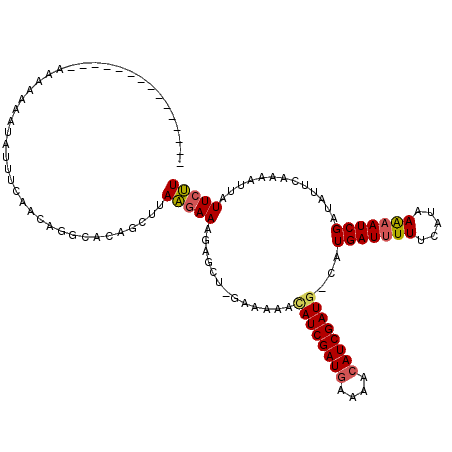
| Location | 502,153 – 502,272 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.94 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -9.84 |
| Energy contribution | -10.28 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 502153 119 + 1281640 UAACAUAAUUUGAAAAAAAAAAUAUUUCAACAGGCACAGCUUAAUAAAGAGCUGGAGAAAUAUCGAUGAAAACAUCGAUG-CAUGAUUUUUCAUAAAAAUCGAUAUUCAACAUUAUUCUU ....((((((((((...................((.((((((......)))))).......(((((((....))))))))-).(((((((.....)))))))...))))).))))).... ( -24.10) >DroSec_CAF1 8426 104 + 1 ---------------AAAAAAAUAUUUUAACAGGCACAGUUUAAGAAAGAGCUAGAAAAACAUCGAUGAAAACAUCGAUG-CAUGAUUUUUCAUAAAAAUCGAUAUUCAAAAUUAUUCUU ---------------......................(((((......))))).(((...((((((((....))))))))-..(((((((.....)))))))...)))............ ( -16.20) >DroSim_CAF1 8581 104 + 1 ---------------AAAAAAAUAUUUUAACAGGCACAGUUUAAGAAAGAGCUCGAAAAACAUCGAUGAAAACAUCGAUG-UAUGAUUUUUCAUAAAAAUCGAUAUUCAAAAUUAUUCUU ---------------......................((..(((....(((.((((...(((((((((....))))))))-)((((....)))).....))))..)))....)))..)). ( -17.80) >DroYak_CAF1 12965 96 + 1 UAACAUUAGUUGAAGAC--GAUGAUUUCAACAGGCACGGCUUAGG----------------AUCGAUAAUAACAUCGAUUCCCUGAUGUUUUGUAAGAAUCGAUGUUCAAC------CCU ........((((((.((--(((..((.(((..(((((((....((----------------((((((......)))))))).))).))))))).))..)))).).))))))------... ( -23.50) >consensus _______________AAAAAAAUAUUUCAACAGGCACAGCUUAAGAAAGAGCU_GAAAAACAUCGAUGAAAACAUCGAUG_CAUGAUUUUUCAUAAAAAUCGAUAUUCAAAAUUAUUCUU ..........................................(((((.............((((((((....))))))))...(((((((.....))))))).............))))) ( -9.84 = -10.28 + 0.44)

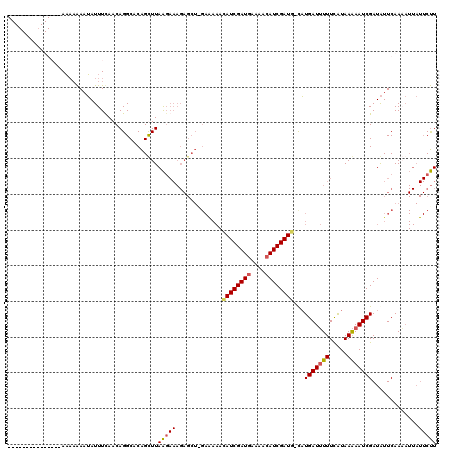
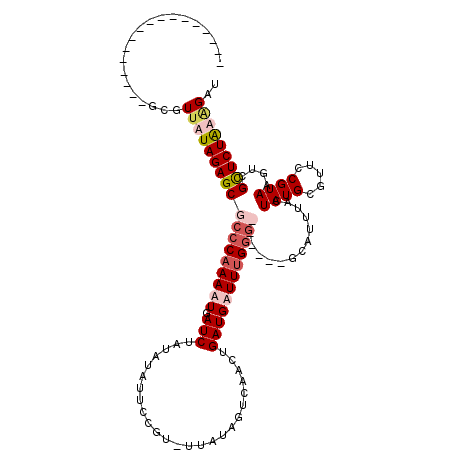
| Location | 502,153 – 502,272 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.94 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -11.56 |
| Energy contribution | -13.12 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 502153 119 - 1281640 AAGAAUAAUGUUGAAUAUCGAUUUUUAUGAAAAAUCAUG-CAUCGAUGUUUUCAUCGAUAUUUCUCCAGCUCUUUAUUAAGCUGUGCCUGUUGAAAUAUUUUUUUUUUCAAAUUAUGUUA ..((..((((((((((((((((...(((((....)))))-.)))))))).....))))))))..))(((((........)))))......((((((.........))))))......... ( -22.70) >DroSec_CAF1 8426 104 - 1 AAGAAUAAUUUUGAAUAUCGAUUUUUAUGAAAAAUCAUG-CAUCGAUGUUUUCAUCGAUGUUUUUCUAGCUCUUUCUUAAACUGUGCCUGUUAAAAUAUUUUUUU--------------- (((((((.((((((.....((((((.....))))))..(-((((((((....))))))))).............................)))))))))))))..--------------- ( -18.60) >DroSim_CAF1 8581 104 - 1 AAGAAUAAUUUUGAAUAUCGAUUUUUAUGAAAAAUCAUA-CAUCGAUGUUUUCAUCGAUGUUUUUCGAGCUCUUUCUUAAACUGUGCCUGUUAAAAUAUUUUUUU--------------- (((((..(.((((((....((((((.....))))))..(-((((((((....)))))))))..)))))).)..)))))...........................--------------- ( -20.30) >DroYak_CAF1 12965 96 - 1 AGG------GUUGAACAUCGAUUCUUACAAAACAUCAGGGAAUCGAUGUUAUUAUCGAU----------------CCUAAGCCGUGCCUGUUGAAAUCAUC--GUCUUCAACUAAUGUUA (((------(((((((((((((((((...........)))))))))))).....)))))----------------)))....(((....((((((......--...))))))..)))... ( -26.50) >consensus AAGAAUAAUGUUGAAUAUCGAUUUUUAUGAAAAAUCAUG_CAUCGAUGUUUUCAUCGAUGUUUUUC_AGCUCUUUCUUAAACUGUGCCUGUUAAAAUAUUUUUUU_______________ ..(((.((((((((((((((((...(((((....)))))..)))))))).....)))))))).)))...................................................... (-11.56 = -13.12 + 1.56)

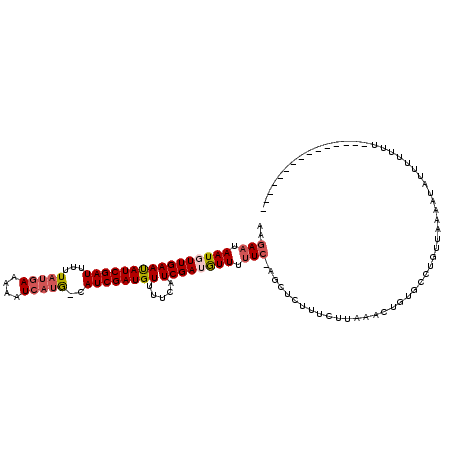
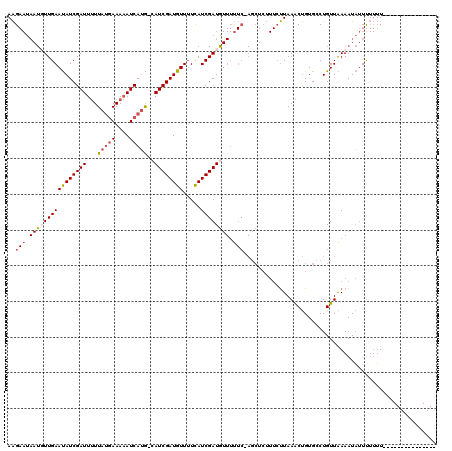
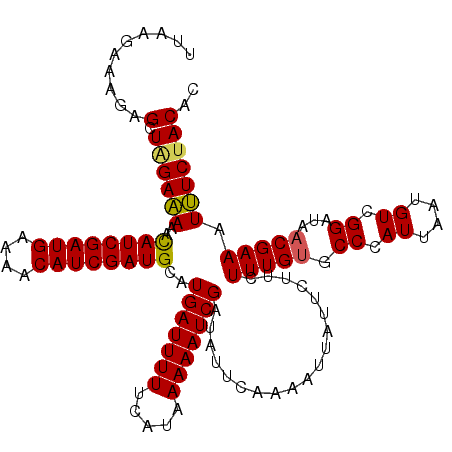
| Location | 502,193 – 502,311 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -18.57 |
| Energy contribution | -18.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 502193 118 + 1281640 UUAAUAAAGAGCUGGAGAAAUAUCGAUGAAAACAUCGAUGCAUGAUUUUUCAUAAAAAUCGAUAUUCAACAUUAUUCUUCUUUGCGCCCAUAUAUGUAGGAUAACGAAAUCUCUACAC ..........((.((((((.((((((((....))))))))..(((((((.....))))))).............))))))...)).........((((((((......))).))))). ( -25.50) >DroSec_CAF1 8451 118 + 1 UUAAGAAAGAGCUAGAAAAACAUCGAUGAAAACAUCGAUGCAUGAUUUUUCAUAAAAAUCGAUAUUCAAAAUUAUUCUUCUUUGUGCCCAUUAAUGUCGGAUAACGAAAUUUCUACAC ...((((((.((.((((...((((((((....))))))))..(((((((.....)))))))................))))....)).).......(((.....)))..))))).... ( -21.40) >DroSim_CAF1 8606 118 + 1 UUAAGAAAGAGCUCGAAAAACAUCGAUGAAAACAUCGAUGUAUGAUUUUUCAUAAAAAUCGAUAUUCAAAAUUAUUCUUCUUUGUGCCCAUUAAUGUCGGAUAACGAAAUUUCUACAC ...((((((.((.((((..(((((((((....))))))))).(((((((.....)))))))...................)))).)).).......(((.....)))..))))).... ( -24.30) >consensus UUAAGAAAGAGCUAGAAAAACAUCGAUGAAAACAUCGAUGCAUGAUUUUUCAUAAAAAUCGAUAUUCAAAAUUAUUCUUCUUUGUGCCCAUUAAUGUCGGAUAACGAAAUUUCUACAC ..........(.((((((..((((((((....))))))))..(((((((.....)))))))...................(((((.((.((....)).))...))))).))))))).. (-18.57 = -18.47 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:46 2006