| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 376,815 – 376,908 |
| Length | 93 |
| Max. P | 0.999937 |

| Location | 376,815 – 376,908 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -20.65 |
| Energy contribution | -20.43 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.81 |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
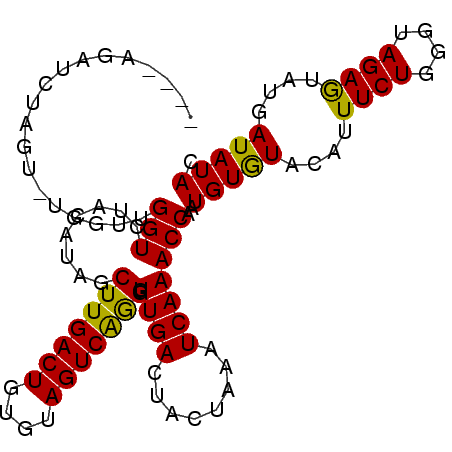
>4_DroMel_CAF1 376815 93 + 1281640 AUAUAUCAUAUUCUAGCAAGAAAGUUACACAAUGGUUUGACUGAGUAGUCAAUCUCGACUACACAGUCAAGCUUUUUACACCCGU--------UUCUCGUU ..............(((.(((((..........((((((((((.((((((......)))))).))))))))))...........)--------)))).))) ( -26.40) >DroSec_CAF1 1496 97 + 1 GUAUAUCAUACUCUACCCAGAAAUGUAUACAUUGGUUUGAUUUAGUAGUCAACCCUGACUACACAGUCAAGCUAUGUAGACCAACCAAACUAGAUCU---- ...........((((.........((.(((((.(((((((((..(((((((....)))))))..)))))))))))))).)).........))))...---- ( -23.77) >DroSim_CAF1 1602 96 + 1 GUAUAUCAUACUCUACCCAGAAAUGUACACAUGGGUUUGAUUUAGUAGUCAACCCUGACUACCCAGUCAAGCUAUGUGGACCAACCA-ACUAGAUCU---- ...........((((..((....))..(((((((.(((((((..(((((((....)))))))..)))))))))))))).........-..))))...---- ( -26.40) >consensus GUAUAUCAUACUCUACCCAGAAAUGUACACAUUGGUUUGAUUUAGUAGUCAACCCUGACUACACAGUCAAGCUAUGUAGACCAACCA_ACUAGAUCU____ ...........................(((((.(((((((((..((((((......))))))..))))))))))))))....................... (-20.65 = -20.43 + -0.22)


| Location | 376,815 – 376,908 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -14.91 |
| Energy contribution | -14.03 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 376815 93 - 1281640 AACGAGAA--------ACGGGUGUAAAAAGCUUGACUGUGUAGUCGAGAUUGACUACUCAGUCAAACCAUUGUGUAACUUUCUUGCUAGAAUAUGAUAUAU ..((((((--------(...(..(((...(.(((((((.(((((((....))))))).))))))).)..)))..)...)))))))................ ( -25.90) >DroSec_CAF1 1496 97 - 1 ----AGAUCUAGUUUGGUUGGUCUACAUAGCUUGACUGUGUAGUCAGGGUUGACUACUAAAUCAAACCAAUGUAUACAUUUCUGGGUAGAGUAUGAUAUAC ----.......((((((((((((..........))))..((((((......))))))..))))))))....((((((((((((....)))).))).))))) ( -21.90) >DroSim_CAF1 1602 96 - 1 ----AGAUCUAGU-UGGUUGGUCCACAUAGCUUGACUGGGUAGUCAGGGUUGACUACUAAAUCAAACCCAUGUGUACAUUUCUGGGUAGAGUAUGAUAUAC ----...((((.(-..(...((.(((((.(.((((...(((((((......)))))))...)))).)..))))).))....)..).))))........... ( -26.20) >consensus ____AGAUCUAGU_UGGUUGGUCUACAUAGCUUGACUGUGUAGUCAGGGUUGACUACUAAAUCAAACCAAUGUGUACAUUUCUGGGUAGAGUAUGAUAUAC ...................(((........(((((((....))))))).((((........)))))))..(((((....((((....))))....))))). (-14.91 = -14.03 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:11 2006