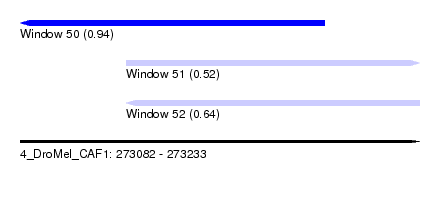
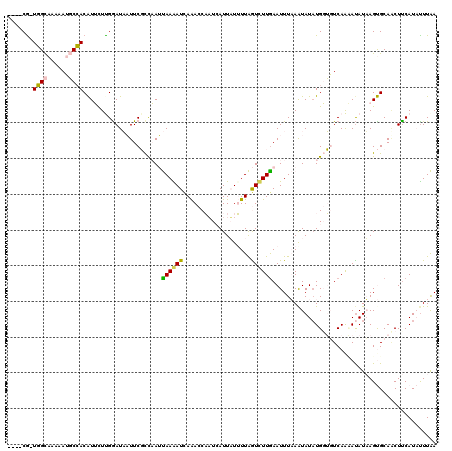
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 273,082 – 273,233 |
| Length | 151 |
| Max. P | 0.940044 |

| Location | 273,082 – 273,197 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.20 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -3.35 |
| Energy contribution | -2.51 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.15 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 273082 115 - 1281640 ----CG-UGGCAAACAUGCCACAUUCUUGAAUAAUUCGCCAAUUAAAAUCAAACCAUUCAUUAUUUUAGUCUUGAAUUUAAAUAUAUGGUGUCAAUACAUUAGUGCAACUUCAUAUUUAA ----.(-(((((....)))))).....((((((...(((((.(((((.((((..(.............)..)))).))))).....)))))....(((....)))........)))))). ( -17.62) >DroPse_CAF1 3578 118 - 1 GCGUCAUUUGU--AAAAUCAACGUGCUUGGAUUUUUCCUUCAACAAAAUUAGACCUAUAAUUUUGUUGAUUUUGAAUUUUAAUAUAUGUGUUCAACAUAUAAGCGCAACUUCAUUUUUGA ((((......(--((((((((.......(((....))).((((((((((((......))))))))))))..))).)))))).(((((((.....))))))).)))).............. ( -27.70) >DroEre_CAF1 2932 115 - 1 ----CG-UGGCAAACAUGCCACAUUUUCGGAUAAUUCGCCAAUUAAGAUCAAACCAAUCAUUAUUUUAGUCUUGAAUUUAAAUAUUUGGUGUCAGAAUAUAAGUGCAUCUUCAUACUUAA ----.(-(((((....))))))...(((.((.....(((((((((((.((((((.((........)).)).)))).)))))....)))))))).)))..((((((........)))))). ( -21.10) >DroYak_CAF1 18376 115 - 1 ----CG-UGGCAAAAAUGCCACAUUUUCGGAUAAUUCGCCAAUUAAGAUCAAACCAUUCAUUAUUUUAGUCUUAAAUUUAAGUAUAUGGUGUCAGAAUAUUAGUGCAGCUCCAUAUUUAA ----.(-(((((....))))))......(((.....(((.(((.........(((((....(((((.(((.....))).))))).)))))........))).)))....)))........ ( -19.93) >DroPer_CAF1 5205 118 - 1 GCGUCAUUUGU--AAAAUCAACGUGCUUGGAUUUUUCCUUCAACAAAAUUAAACCUAUAAUUUUGUUGAUUUUGAAUUUUAAUAUAUGUGUUCAACAUAUAAGCGCAACUUCAUUUUUGA ((((......(--((((((((.......(((....))).((((((((((((......))))))))))))..))).)))))).(((((((.....))))))).)))).............. ( -26.90) >consensus ____CG_UGGCAAAAAUGCCACAUUCUUGGAUAAUUCGCCAAUUAAAAUCAAACCAAUCAUUAUUUUAGUCUUGAAUUUAAAUAUAUGGUGUCAAAAUAUAAGUGCAACUUCAUAUUUAA .......((((......))))......................((((((...................)))))).............................................. ( -3.35 = -2.51 + -0.84)

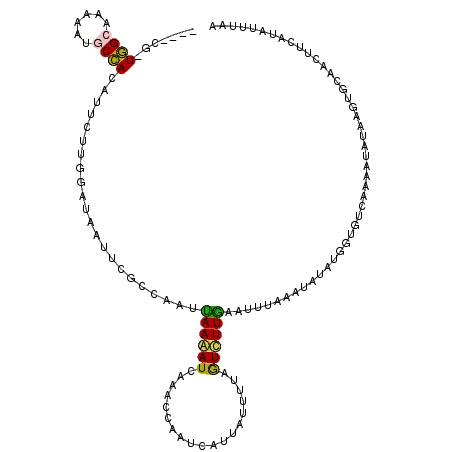
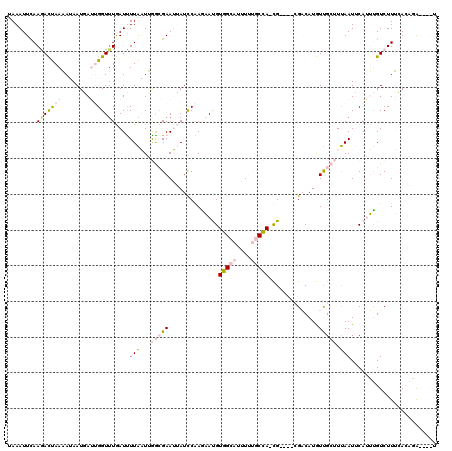
| Location | 273,122 – 273,233 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.08 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -4.59 |
| Energy contribution | -5.79 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.19 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 273122 111 + 1281640 UAAAUUCAAGACUAAAAUAAUGAAUGGUUUGAUUUUAAUUGGCGAAUUAUUCAAGAAUGUGGCAUGUUUGCCA-CG----CGACAUGUUGCUUUAAUUCAUUUGUCUUUCACAGA----U .......(((((........(((((((((((...........))))))))))).((((((((((....)))))-)(----(((....))))....))))....))))).......----. ( -28.40) >DroPse_CAF1 3618 113 + 1 AAAAUUCAAAAUCAACAAAAUUAUAGGUCUAAUUUUGUUGAAGGAAAAAUCCAAGCACGUUGAUUUU--ACAAAUGACGCCGCCAUAUCGG-UCAAUACAAGCGUCUUGAAAUUG----C .(((.((((..((((((((((((......)))))))))))).(((....))).......)))).)))--.(((..(((((.(((.....))-)........))))))))......----. ( -23.20) >DroEre_CAF1 2972 115 + 1 UAAAUUCAAGACUAAAAUAAUGAUUGGUUUGAUCUUAAUUGGCGAAUUAUCCGAAAAUGUGGCAUGUUUGCCA-CG----AGACAUGUUGCUUUAAUUCAUUUGUCUUUCACAGAUACAU ........(((((((........))))))).((((.....(((((((.....(((...(..((((((((....-..----))))))))..).....))))))))))......)))).... ( -21.70) >DroYak_CAF1 18416 111 + 1 UAAAUUUAAGACUAAAAUAAUGAAUGGUUUGAUCUUAAUUGGCGAAUUAUCCGAAAAUGUGGCAUUUUUGCCA-CG----AGACAUGUUGCUUUAAUUCAUUUGUCUUUCACAGA----U .......(((((......((((((((((.((.((((..((((........))))....((((((....)))))-))----)))))....)))...))))))).))))).......----. ( -23.00) >DroPer_CAF1 5245 113 + 1 AAAAUUCAAAAUCAACAAAAUUAUAGGUUUAAUUUUGUUGAAGGAAAAAUCCAAGCACGUUGAUUUU--ACAAAUGACGCCGCCAUAUCGG-UCAAUACAAGCGUCUUGAAAUUG----C .(((.((((..((((((((((((......)))))))))))).(((....))).......)))).)))--.(((..(((((.(((.....))-)........))))))))......----. ( -24.00) >consensus UAAAUUCAAGACUAAAAUAAUGAUUGGUUUGAUUUUAAUUGGCGAAUUAUCCAAGAAUGUGGCAUUUUUGCCA_CG____CGACAUGUUGCUUUAAUUCAUUUGUCUUUCACAGA____U ........(((((((........)))))))....(((...(((((..............(((((....)))))..............))))).)))........................ ( -4.59 = -5.79 + 1.20)

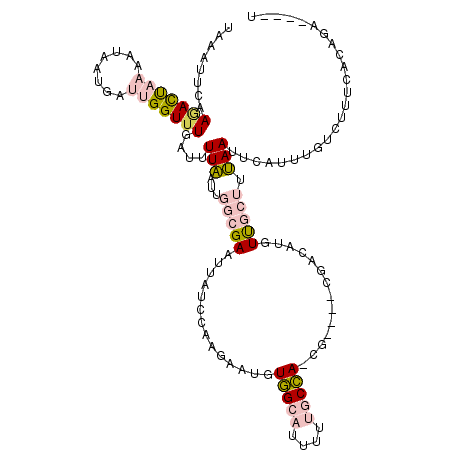
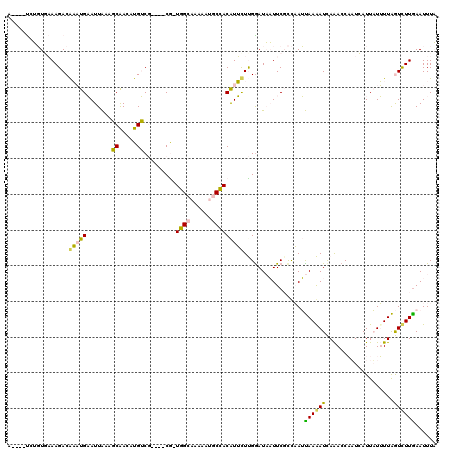
| Location | 273,122 – 273,233 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.08 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -5.11 |
| Energy contribution | -3.35 |
| Covariance contribution | -1.76 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
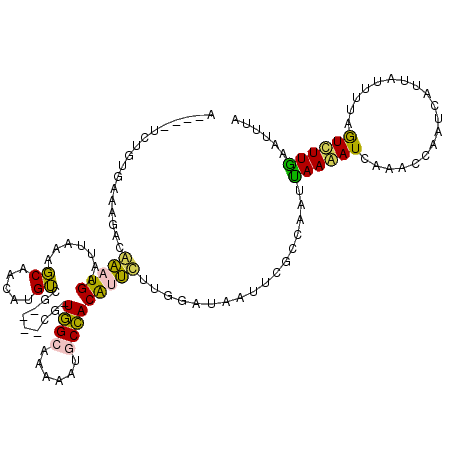
>4_DroMel_CAF1 273122 111 - 1281640 A----UCUGUGAAAGACAAAUGAAUUAAAGCAACAUGUCG----CG-UGGCAAACAUGCCACAUUCUUGAAUAAUUCGCCAAUUAAAAUCAAACCAUUCAUUAUUUUAGUCUUGAAUUUA .----.......(((((....((((((...(((.((((.(----((-((.....))))).))))..)))..))))))......((((((.............)))))))))))....... ( -19.62) >DroPse_CAF1 3618 113 - 1 G----CAAUUUCAAGACGCUUGUAUUGA-CCGAUAUGGCGGCGUCAUUUGU--AAAAUCAACGUGCUUGGAUUUUUCCUUCAACAAAAUUAGACCUAUAAUUUUGUUGAUUUUGAAUUUU .----.........((((((((((((..-..))))))..)))))).....(--(((((((((((....(((....)))....))(((((((......)))))))))))))))))...... ( -27.40) >DroEre_CAF1 2972 115 - 1 AUGUAUCUGUGAAAGACAAAUGAAUUAAAGCAACAUGUCU----CG-UGGCAAACAUGCCACAUUUUCGGAUAAUUCGCCAAUUAAGAUCAAACCAAUCAUUAUUUUAGUCUUGAAUUUA ...((((((.((((((((..((........))...)))))----.(-(((((....)))))).))).)))))).........(((((((.(((...........))).)))))))..... ( -21.90) >DroYak_CAF1 18416 111 - 1 A----UCUGUGAAAGACAAAUGAAUUAAAGCAACAUGUCU----CG-UGGCAAAAAUGCCACAUUUUCGGAUAAUUCGCCAAUUAAGAUCAAACCAUUCAUUAUUUUAGUCUUAAAUUUA (----((((.((((((((..((........))...)))))----.(-(((((....)))))).))).)))))..........(((((((.(((...........))).)))))))..... ( -20.20) >DroPer_CAF1 5245 113 - 1 G----CAAUUUCAAGACGCUUGUAUUGA-CCGAUAUGGCGGCGUCAUUUGU--AAAAUCAACGUGCUUGGAUUUUUCCUUCAACAAAAUUAAACCUAUAAUUUUGUUGAUUUUGAAUUUU .----.........((((((((((((..-..))))))..)))))).....(--(((((((((((....(((....)))....))(((((((......)))))))))))))))))...... ( -26.60) >consensus A____UCUGUGAAAGACAAAUGAAUUAAAGCAACAUGUCG____CG_UGGCAAAAAUGCCACAUUCUUGGAUAAUUCGCCAAUUAAAAUCAAACCAAUCAUUAUUUUAGUCUUGAAUUUA .................(((((.......((.....)).........((((......))))))))).................((((((...................))))))...... ( -5.11 = -3.35 + -1.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:58 2006