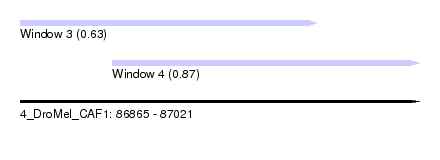
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 86,865 – 87,021 |
| Length | 156 |
| Max. P | 0.868040 |

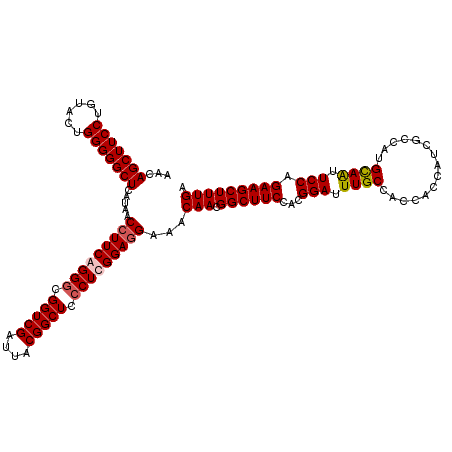
| Location | 86,865 – 86,981 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.48 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -27.85 |
| Energy contribution | -28.85 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 86865 116 + 1281640 AAUACAUAAAA---AAAUAUGCAUUUCAUAUUUAAUUUAAACAGCUUCCUGUACUGGGGGCUCAUAACCUUCAGGGCGGUCGAUUACGGCUCCCUCGGAUGAAACAACGGCUUCCACGG ....((((...---...))))..((((((.............(((((((......))))))).....((...((((.(((((....))))))))).))))))))...((.......)). ( -26.00) >DroSec_CAF1 29164 116 + 1 AAUACAUUAAA---AAAUAUGCAUUUCAUAUUUAAUUUAAACAGCUUCCUGUACUGGGGGCUCAUAACCUUCAGGGCGGUCGAUUACGGCUCCCUCGGAGGAAACAACGGCUUCCACGG ......(((((---(((((((.....)))))))..)))))((((....))))..((((((((.....(((((((((.(((((....))))))))).))))).......))))))))... ( -32.70) >DroEre_CAF1 46379 118 + 1 AAUACAUUUAAAAAAAAUACGCAUUUCAUAUUCAAU-UAAACAGCUUCCUGUACGGGGGGCUCAUAACCUUCGGGACGGUCGAUUACGGCUCCCUCGGAGGAAACAACGGCUUCCACGG ....................................-...((((....)))).(((((((((.....((((((((..(((((....)))))..)))))))).......))))))).)). ( -30.60) >DroYak_CAF1 52556 116 + 1 AAUACAUUGCA---AAAUAUGCAUUUCAUAUUCAAUUUAAACAGCUUCCUGUACUGGGGGCUCAUAACCUUCGGGGCGGUCGAUCACGGCUCCCUCGGAGGAAACAACGGCUUCCACGG .......((((---.....)))).................((((....))))..((((((((.....(((((((((.(((((....))))).))))))))).......))))))))... ( -35.70) >consensus AAUACAUUAAA___AAAUAUGCAUUUCAUAUUCAAUUUAAACAGCUUCCUGUACUGGGGGCUCAUAACCUUCAGGGCGGUCGAUUACGGCUCCCUCGGAGGAAACAACGGCUUCCACGG ....................((....................(((((((......))))))).....(((((((((.(((((....))))).)))))))))........))........ (-27.85 = -28.85 + 1.00)

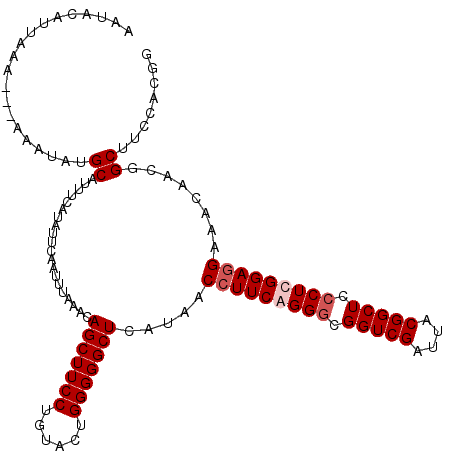
| Location | 86,901 – 87,021 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -42.49 |
| Consensus MFE | -39.53 |
| Energy contribution | -40.04 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 86901 120 + 1281640 AACAGCUUCCUGUACUGGGGGCUCAUAACCUUCAGGGCGGUCGAUUACGGCUCCCUCGGAUGAAACAACGGCUUCCACGGAUUUGCCACCGCCAUCGCCAUGCAAUUCCAGAAGCUUUGA ....((((((......))))))((((..((...((((.(((((....))))))))).))))))..(((.((((((...(((.((((....((....))...)))).))).))))))))). ( -38.30) >DroSec_CAF1 29200 120 + 1 AACAGCUUCCUGUACUGGGGGCUCAUAACCUUCAGGGCGGUCGAUUACGGCUCCCUCGGAGGAAACAACGGCUUCCACGGAUUUGCCACCGCCAUCGCCAUGCAAUUCCAGAAGCUUUGA ...(((((((......))))))).....(((((((((.(((((....))))))))).)))))...(((.((((((...(((.((((....((....))...)))).))).))))))))). ( -44.40) >DroEre_CAF1 46417 120 + 1 AACAGCUUCCUGUACGGGGGGCUCAUAACCUUCGGGACGGUCGAUUACGGCUCCCUCGGAGGAAACAACGGCUUCCACGGAUUUGCCACCACCAUCGCCAUGUAGUUCCAGAAGCUUUGA ...((((((((....))(((((((((..((((((((..(((((....)))))..)))))))).......(((.((....))...)))............))).)))))).)))))).... ( -40.20) >DroYak_CAF1 52592 120 + 1 AACAGCUUCCUGUACUGGGGGCUCAUAACCUUCGGGGCGGUCGAUCACGGCUCCCUCGGAGGAAACAACGGCUUCCACGGAUUUGCCACCACCAUCGCCAUGUAGAUCCAGAAGCUUUGA ...(((((((......))))))).....(((((((((.(((((....))))).)))))))))...(((.((((((...((((((((...............)))))))).))))))))). ( -47.06) >consensus AACAGCUUCCUGUACUGGGGGCUCAUAACCUUCAGGGCGGUCGAUUACGGCUCCCUCGGAGGAAACAACGGCUUCCACGGAUUUGCCACCACCAUCGCCAUGCAAUUCCAGAAGCUUUGA ...(((((((......))))))).....(((((((((.(((((....))))).)))))))))...(((.((((((...(((.((((...............)))).))).))))))))). (-39.53 = -40.04 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:49 2006