| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 263,492 – 263,599 |
| Length | 107 |
| Max. P | 0.914361 |

| Location | 263,492 – 263,599 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.86 |
| Mean single sequence MFE | -33.31 |
| Consensus MFE | -13.97 |
| Energy contribution | -15.75 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
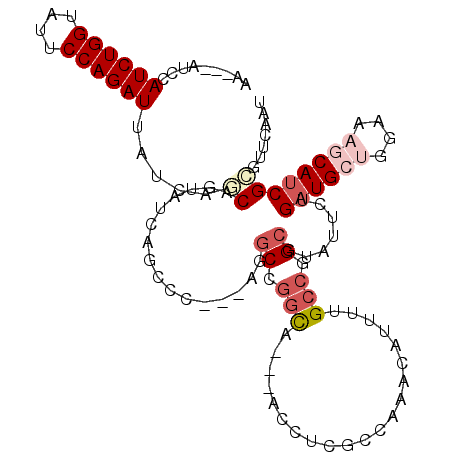
>4_DroMel_CAF1 263492 107 + 1281640 AUAUCAUCCAUCUGGUAUUCCAGAUUUUCAGCAGUAUCAACAA---AGGCCUGUU---CUAAGUCCAAACAUUUUGCCGGCGUAUUCUGAUGCUGGAAAGCACCGCGUCCAAU .........((((((....))))))((((..((((((((....---(.(((.((.---..((((......)))).)).))).)....)))))))))))).............. ( -23.00) >DroVir_CAF1 7513 104 + 1 CA---GUCCAUCUGGUAUUCCAGAUUACCAACAAUAUCAGGCA---AGGCCGGCG---ACCUCGCCGAGCAUAUUGCCAGCUUAUUCGGAUGCUGGAAAGCAUCGGGUUCAAC ..---....((((((....))))))..((..........((((---(.(((((((---....))))).))...)))))..........((((((....))))))))....... ( -40.70) >DroPse_CAF1 3526 107 + 1 AAAUCAUCCAUCUGGUAUUCCAGAUUAUCAGCAGUAUCAGUCC---AAGCCGGCA---ACCUCGCCAAACAUUUUGCCGGCGUAUUCUGAUGCGGGAAAACAUCGCGUUCAAU ......(((((((((....))))))........(((((((...---..(((((((---(..............)))))))).....))))))).)))................ ( -34.74) >DroWil_CAF1 3762 104 + 1 CA---GUCCAUCUGGUAUUCCAGAUUACCAGCAGUAUCAGCCG---AGGCCGAUC---UCUUCACCGAACAUUGUGCCCGCCUAUUCAGAUGCUGGCAAGCAUCGGGUGCAAU ..---((.(((((((....))))))....((.((.(((.((..---..)).))))---))).....).))((((..((((......).((((((....)))))))))..)))) ( -31.00) >DroMoj_CAF1 4844 110 + 1 CA---GUCCAUCUGGUAUUCCAGAUUAUCAACAAUAUCAGGUCAAUAGACCGGCUGCUGCAUCACCAAGCAUCGUGCCAGCGUAUUCCGAUGCUGGCAAGCAUCGUGUCCAAU ((---(((.((((((....))))))..............((((....))))))))).((...(((...((....(((((((((......))))))))).))...)))..)).. ( -35.70) >DroPer_CAF1 3597 107 + 1 AAAUCAUCCAUCUGGUAUUCCAGAUUAUCAGCAGUAUCAGUCC---AAGCCGGCA---ACCUCGCCAAACAUUUUGCCGGCGUAUUCUGAUGCGGGAAAACAUCGCGUUCAAU ......(((((((((....))))))........(((((((...---..(((((((---(..............)))))))).....))))))).)))................ ( -34.74) >consensus AA___AUCCAUCUGGUAUUCCAGAUUAUCAGCAGUAUCAGCCC___AGGCCGGCA___ACCUCGCCAAACAUUUUGCCGGCGUAUUCUGAUGCUGGAAAGCAUCGCGUUCAAU .........((((((....)))))).....((................((.(((.....................))).)).......((((((....))))))))....... (-13.97 = -15.75 + 1.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:55 2006