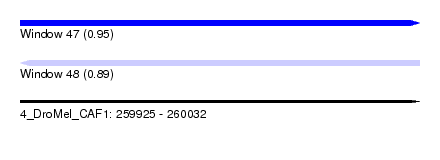
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 259,925 – 260,032 |
| Length | 107 |
| Max. P | 0.954810 |

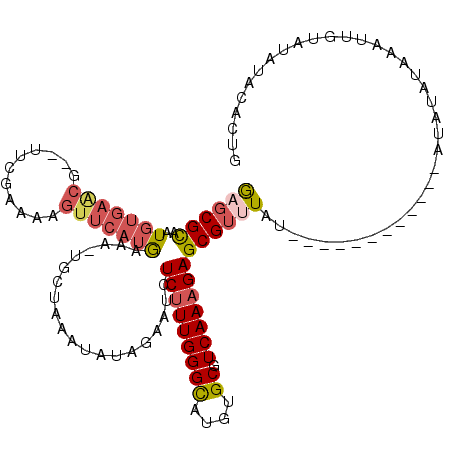
| Location | 259,925 – 260,032 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -10.68 |
| Energy contribution | -10.84 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
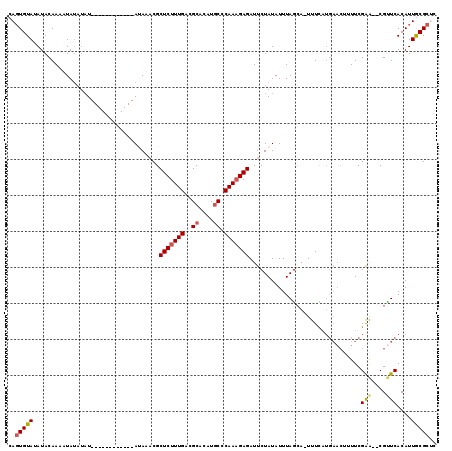
>4_DroMel_CAF1 259925 107 + 1281640 CAGUGUAUAUACAAGUUAUAUAU------------ACAAUCGCUCUUUGACGCACAUGCCCAAAGAGAUUCUAUAUUUAGCAAUUUCAUGAACUUUUCGAACACGUUCACAUUGCGCUC ..(((((((((.....)))))))------------)).....(((((((..((....)).)))))))............(((((....(((((...........))))).))))).... ( -23.50) >DroSec_CAF1 2207 100 + 1 CAGUGUAUAUACAAUAUA----U------------AUAUACGCUCUUUGACGCACAUGCCCAAAGAGAUUCUAUAUUUAACA-UUUUAUGAACUUUUCGAA--CGUUCACAUUGCGCGC ..(((((((((.....))----)------------)))))).(((((((..((....)).)))))))...............-.....(((((........--.))))).......... ( -19.20) >DroSim_CAF1 2157 104 + 1 CAGUGUAUAUACAAUAUAUGUAU------------AUAUACGCUCUUUGACGCACAUGCCCAAAGAGAUUCUAUAUUUAGCA-UUUUAUGAACUUUUCGAA--CGUUCACAUUGCGCGC ..(((((((((((.....)))))------------)))))).(((((((..((....)).)))))))............(((-.....(((((........--.)))))...))).... ( -26.80) >DroEre_CAF1 1494 111 + 1 CAUUGUACAUACAAAUUAUAUAUAUGUAUGUACAUAUAAACGCUCUUUGAGGCACAUGCCCAAAGAGAUUCUAUAUUUAGCAUUUUC------AUUUCGAG--CGCUCACAUUACGCUU ...((((((((((.((.....)).))))))))))((((....(((((((.(((....))))))))))....))))............------.....(((--((.........))))) ( -30.30) >DroYak_CAF1 1703 105 + 1 CAGUGUACAUACAAAGUAUAUAU------------AAAAACGCUCUUUGAAGCACAUACCCAAUGAGAUUCUAUAUUUAGAAGUUUCAUGAACAUUUCGGA--CGCUCACAUUACGCUC .((((((....(((((.......------------.........))))).(((.........(((((((((((....))).))))))))((.....))...--.))).....)))))). ( -16.19) >consensus CAGUGUAUAUACAAAAUAUAUAU____________AUAAACGCUCUUUGACGCACAUGCCCAAAGAGAUUCUAUAUUUAGCA_UUUCAUGAACUUUUCGAA__CGUUCACAUUGCGCUC ..(((((...................................(((((((..((....)).)))))))...............................(((....)))....))))).. (-10.68 = -10.84 + 0.16)

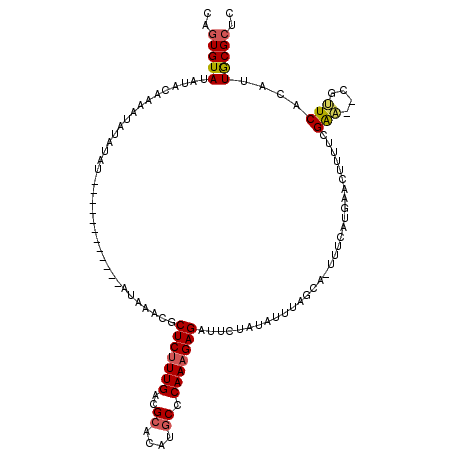
| Location | 259,925 – 260,032 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -14.30 |
| Energy contribution | -15.38 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 259925 107 - 1281640 GAGCGCAAUGUGAACGUGUUCGAAAAGUUCAUGAAAUUGCUAAAUAUAGAAUCUCUUUGGGCAUGUGCGUCAAAGAGCGAUUGU------------AUAUAUAACUUGUAUAUACACUG ..(((((((((((((...........))))))...))))).............(((((((((....)).)))))))))...(((------------((((((.....)))))))))... ( -27.10) >DroSec_CAF1 2207 100 - 1 GCGCGCAAUGUGAACG--UUCGAAAAGUUCAUAAAA-UGUUAAAUAUAGAAUCUCUUUGGGCAUGUGCGUCAAAGAGCGUAUAU------------A----UAUAUUGUAUAUACACUG ((((((..(((((((.--........)))))))..(-((((......((.....))...))))))))))).....((.((((((------------(----(.....)))))))).)). ( -24.80) >DroSim_CAF1 2157 104 - 1 GCGCGCAAUGUGAACG--UUCGAAAAGUUCAUAAAA-UGCUAAAUAUAGAAUCUCUUUGGGCAUGUGCGUCAAAGAGCGUAUAU------------AUACAUAUAUUGUAUAUACACUG (((((((.(((((((.--........)))))))...-))).............(((((((((....)).))))))))))).(((------------(((((.....))))))))..... ( -28.90) >DroEre_CAF1 1494 111 - 1 AAGCGUAAUGUGAGCG--CUCGAAAU------GAAAAUGCUAAAUAUAGAAUCUCUUUGGGCAUGUGCCUCAAAGAGCGUUUAUAUGUACAUACAUAUAUAUAAUUUGUAUGUACAAUG .(((((.......)))--))......------...........(((((((..((((((((((....))).)))))))..)))))))(((((((((...........))))))))).... ( -34.00) >DroYak_CAF1 1703 105 - 1 GAGCGUAAUGUGAGCG--UCCGAAAUGUUCAUGAAACUUCUAAAUAUAGAAUCUCAUUGGGUAUGUGCUUCAAAGAGCGUUUUU------------AUAUAUACUUUGUAUGUACACUG ..((.(((((((((((--(.....)))))))......(((((....)))))...))))).)).(((((..(((((.(..(....------------..)..).)))))...)))))... ( -19.50) >consensus GAGCGCAAUGUGAACG__UUCGAAAAGUUCAUGAAA_UGCUAAAUAUAGAAUCUCUUUGGGCAUGUGCGUCAAAGAGCGUUUAU____________AUAUAUAAAUUGUAUAUACACUG ((((((..(((((((...........)))))))....................(((((((((....)).)))))))))))))..................................... (-14.30 = -15.38 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:54 2006