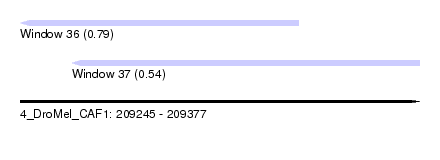
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 209,245 – 209,377 |
| Length | 132 |
| Max. P | 0.793777 |

| Location | 209,245 – 209,337 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -17.87 |
| Consensus MFE | -9.70 |
| Energy contribution | -10.72 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 209245 92 - 1281640 UAGUGCUAUCCUAGCACUUACCAUUAAAGUUACUAUCCCUUACGUAAAAAACGUAAAUACUACGUCUAGUGUAAGGACCCAAAAA-GGAAAAA- .(((((((...)))))))...................(((((((......(((((.....)))))....)))))))..((.....-)).....- ( -18.10) >DroSec_CAF1 3320 92 - 1 UAGUGCUAUACUAGCACUUACCAUUAAAGUUACUAUCCCUUACGUAAGAAACGUAAAUACUACGUGUAGUGUACGGACCCAAAAA-GGAAAAA- .(((((((...))))))).................(((.((((((.....))))))((((((....))))))..))).((.....-)).....- ( -18.60) >DroSim_CAF1 3664 92 - 1 UAGUGCUAUACUAGCACUUACCAUUAAAGUUACUAUCCCUUACGUAAGAAACGUAAAUACUACGUGUAGUGUACGGACCCAAAAA-GGAAAAA- .(((((((...))))))).................(((.((((((.....))))))((((((....))))))..))).((.....-)).....- ( -18.60) >DroEre_CAF1 2664 92 - 1 UAGUGCUAUACUAGCACUCACCAUAAAGGUUACCAUUCCUUACGUAAAAAACGUUCAUACUACGUAUAGUGUAAGGACCCAAAAA-GGAAAAC- .(((((((...))))))).(((.....)))......((((((((......((((.......))))....)))))))).((.....-)).....- ( -19.30) >DroYak_CAF1 1003 92 - 1 UAGUGCUAUACUAGCACUUACCAUUAAGGUUACCAUUCCUUACGUUAAAAACGUGCACACUACGUCUAGUGUAGGGGCCCAAAAA-GGAAAAA- .(((((((...))))))).(((.....))).....((((((((((.....))))((...(((((.....)))))..)).....))-))))...- ( -21.90) >DroMoj_CAF1 4991 84 - 1 UACUGCCCUAUUUGCAUUAACCGUUAAGGUAACUGUACCAUAUAUCCGAAAUGCCC------CAUGCA---UUA-AACACAAAAAUGAAAAUGC .............(((((...((((..((((....))))..........(((((..------...)))---)).-........))))..))))) ( -10.70) >consensus UAGUGCUAUACUAGCACUUACCAUUAAAGUUACUAUCCCUUACGUAAAAAACGUAAAUACUACGUGUAGUGUAAGGACCCAAAAA_GGAAAAA_ .((((((.....))))))..((..................(((((.....))))).((((((....))))))..............))...... ( -9.70 = -10.72 + 1.02)

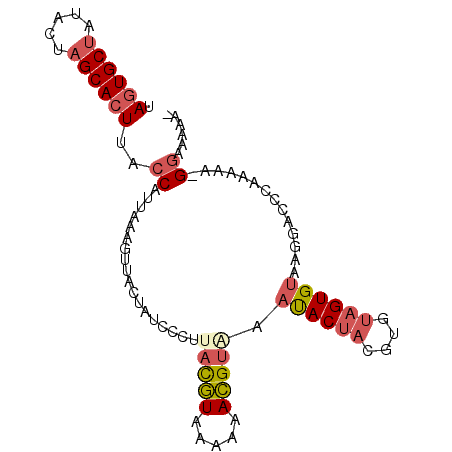
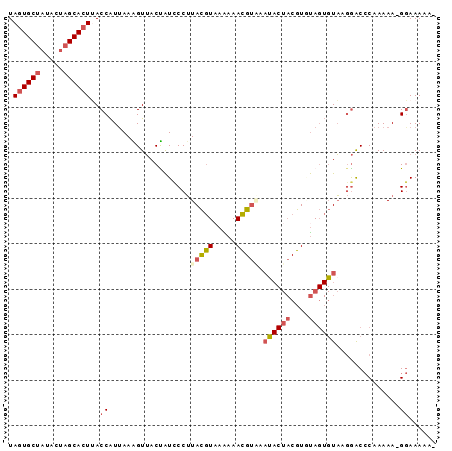
| Location | 209,262 – 209,377 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.20 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 209262 115 - 1281640 AUGGGUUUACAAAACACGCUUUUAGCUAUGCUCUAGUAACUAGUGCUAUCCUAGCACUUACCAUUAAAGUUACUAUCCCUUACGUAAAAAAC--GUAAAUACUACGUCUAGUGUAAG- ..(((............((.....)).......(((((((((((((((...))))))).........)))))))).)))((((((.....))--))))((((((....))))))...- ( -25.80) >DroSec_CAF1 3337 115 - 1 AUGGCUUUACGAAACACGCUUUUAGCUAUGCUCUAGUAACUAGUGCUAUACUAGCACUUACCAUUAAAGUUACUAUCCCUUACGUAAGAAAC--GUAAAUACUACGUGUAGUGUACG- ((((((...((.....)).....))))))....(((((((((((((((...))))))).........))))))))....((((((.....))--))))((((((....))))))...- ( -26.70) >DroSim_CAF1 3681 115 - 1 AUGGGUUUACGAAACACGCUUUUAGCUAUGCUCUAGUAACUAGUGCUAUACUAGCACUUACCAUUAAAGUUACUAUCCCUUACGUAAGAAAC--GUAAAUACUACGUGUAGUGUACG- ..(((............((.....)).......(((((((((((((((...))))))).........)))))))).)))((((((.....))--))))((((((....))))))...- ( -27.10) >DroEre_CAF1 2681 115 - 1 AUGGAAUUACGAAACAUGCUUUAAGCUAUGCUCUUGUAACUAGUGCUAUACUAGCACUCACCAUAAAGGUUACCAUUCCUUACGUAAAAAAC--GUUCAUACUACGUAUAGUGUAAG- ((((..((((((..(((((.....)).)))...))))))..(((((((...)))))))..)))).((((........))))((((.....))--))..((((((....))))))...- ( -23.10) >DroYak_CAF1 1020 115 - 1 AUGGGUUUACAAAAAACGUUUUUAGCUAUGCUAUUGUAACUAGUGCUAUACUAGCACUUACCAUUAAGGUUACCAUUCCUUACGUUAAAAAC--GUGCACACUACGUCUAGUGUAGG- ...............(((((((((((.........(((...(((((((...))))))))))...(((((........))))).)))))))))--))..((((((....))))))...- ( -28.00) >DroPer_CAF1 3451 117 - 1 AUGGAAUUACAAAGCACGCUUUUAGCUGCGUGCUCGUGACCAGUGCGAUAGUAGCUCUUACCAUUAAGAUUACUACUCCGUAUGUAAGAAAUACGCAAUCACAAUCCCCAA-GAAAGA .(((........(((((((........))))))).((((....(((((((((((.(((((....))))))))))).)).((((.......))))))).)))).....))).-...... ( -32.00) >consensus AUGGGUUUACAAAACACGCUUUUAGCUAUGCUCUAGUAACUAGUGCUAUACUAGCACUUACCAUUAAAGUUACUAUCCCUUACGUAAAAAAC__GUAAAUACUACGUCUAGUGUAAG_ ..(((............((.....)).........(((((((((((((...))))))).........))))))...)))...................((((((....)))))).... (-15.36 = -15.20 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:38 2006