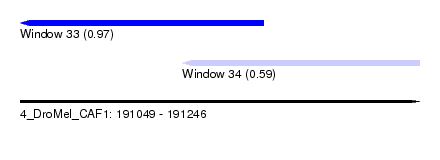
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 191,049 – 191,246 |
| Length | 197 |
| Max. P | 0.967384 |

| Location | 191,049 – 191,169 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -18.97 |
| Energy contribution | -19.53 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 191049 120 - 1281640 AGGUAUCAAGUCAGUCGACCGCGACUGAUAGCGAUACCGAGAAUGAUCGGAAUUGUAAGUCUGAAAGUUCCUUGUUUUCCACUUUGGAAAAUCAAAUUUAUCAAUCUGACAUUACCACGU .((((((..((((((((....))))))))...))))))(..((((.(((((.((((((((.(((...((((..((.....))...))))..))).))))).))))))))))))..).... ( -37.40) >DroSec_CAF1 18474 120 - 1 AGGUAUCAAGUCAGUCGAUCGCGACUGAUAGCGAUACGCAGAUUGAUCAGAAUUCGAAGUCCGAAAGUUCCUUAAUUUCCACUUUGAAAAAUCCAAUUUUUCAAUCUGACAUUACCACGU ..(((((..((((((((....))))))))...)))))(((((((((...((.((((((((..((((.........)))).))))))))...)).......)))))))).).......... ( -31.70) >DroEre_CAF1 20401 120 - 1 UGGUAUCAAGUCUGUCAGCCGCAACUGAUAGCAAAACCCAGAAUAAUCAGAGUUCCGAAUCCGAAGGAUCAUUGAUAUCUACUUUGAAAGACCCAAACUUUCAAUCUGACAUUACCACAU (((((....(.(((((((......))))))))......((((...(((((.(.(((.........))).).))))).......(((((((.......)))))))))))....)))))... ( -27.80) >consensus AGGUAUCAAGUCAGUCGACCGCGACUGAUAGCGAUACCCAGAAUGAUCAGAAUUCCAAGUCCGAAAGUUCCUUGAUUUCCACUUUGAAAAAUCCAAUUUUUCAAUCUGACAUUACCACGU .((((....((((((((....)))))))).................(((((..........(....)................((((((((.....)))))))))))))...)))).... (-18.97 = -19.53 + 0.56)

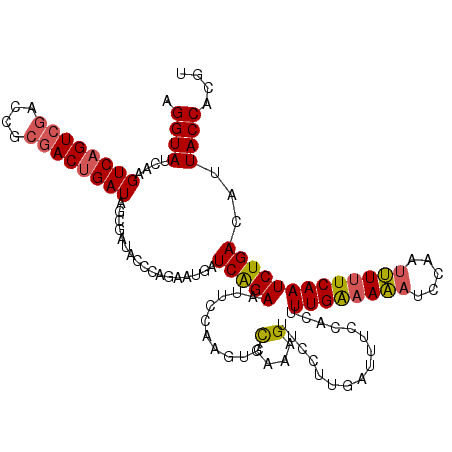
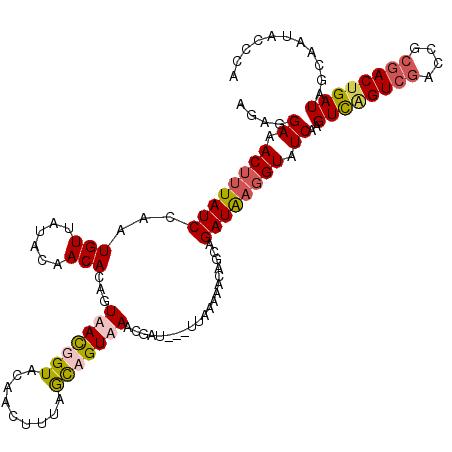
| Location | 191,129 – 191,246 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -16.34 |
| Energy contribution | -17.52 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 191129 117 - 1281640 AGAGGAAACUUUAUCCAAUGUUAUACAACACAGUAACGGUACAACUUUAACAGUAAACGAU---UUAAAAACUGCCGAUAAGGUAUCAAGUCAGUCGACCGCGACUGAUAGCGAUACCGA .(((....)))((((....(((((........)))))((((....(((((..(....)...---)))))...)))))))).((((((..((((((((....))))))))...)))))).. ( -30.30) >DroSec_CAF1 18554 117 - 1 AGAGGAAACUUUAUCCAAUGUUAUACAACACAGUAACGGUACAACUUCAGUAGUAAACGAU---UUAAAAACGGCUGAUAAGGUAUCAAGUCAGUCGAUCGCGACUGAUAGCGAUACGCA ((((....)))).......(((((........)))))(((((....(((((.((.......---......)).)))))....)))))..((((((((....)))))))).(((...))). ( -30.72) >DroEre_CAF1 20481 117 - 1 GGAGGAAACUUUAUCUAAUGUCAUACAACACAGUAACGGUACAACUUUAGCCGUUAACGAU---UUAAAAACAGCAGAUAUGGUAUCAAGUCUGUCAGCCGCAACUGAUAGCAAAACCCA ((..((.(((.(((((..(((............(((((((.........))))))).....---......)))..))))).))).))..(.(((((((......)))))))).....)). ( -27.30) >DroYak_CAF1 17630 120 - 1 AGAGGAAACGUUAUCCAAUGUAAUACAACACAGUAAUGGUACAACUUUGGCCGUUAACGAUAAGUCAUCAUUAGCAGAUGAGGUGUCAAGUUGGUCGACCGCGACUGAUAGCAAUACCCA ...(....)((((((..........((((....(((((((.((....)))))))))..((((..(((((.......)))))..))))..))))((((....)))).))))))........ ( -31.40) >consensus AGAGGAAACUUUAUCCAAUGUUAUACAACACAGUAACGGUACAACUUUAGCAGUAAACGAU___UUAAAAACAGCAGAUAAGGUAUCAAGUCAGUCGACCGCGACUGAUAGCAAUACCCA ....((.((((((((...(((......)))...(((((((.........)))))))....................)))))))).))..((((((((....))))))))........... (-16.34 = -17.52 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:34 2006