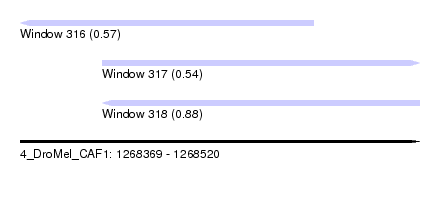
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 1,268,369 – 1,268,520 |
| Length | 151 |
| Max. P | 0.875207 |

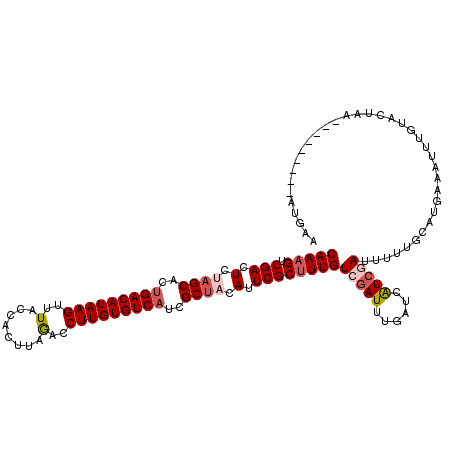
| Location | 1,268,369 – 1,268,480 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -19.44 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1268369 111 - 1281640 CAAAGUCGACUCUAGCACAGACAUAAGUUUACCGCUUAGACCUUGUGUCAUAGCUACAUUCGCUUUGUCGAUUUGGUCAUCAAUUUUCGCAUGAAACUUGUACUAA---------AUGGA (((((.(((.(.((((...((((((((((((.....)))).))))))))...)))).).))))))))((.(((((((...(((.((((....)))).))).)))))---------)).)) ( -30.30) >DroVir_CAF1 12000 120 - 1 CAAAGUCGACUCUAGCACUGACAUAAGUUUGCCACUUAAUCCUUGUGUCAUCGCUACAUUCGCUUUGUCGAUUUGAUCAUCGAUUUUUGCGUGGAAUUUGUACUGUUUUUUUAUUUUGAU ((((.((.((..((((..(((((((((..(........)..)))))))))..)))).....((...((((((......))))))....)))).)).)))).................... ( -28.10) >DroGri_CAF1 14130 111 - 1 CAAAGUCGACUCCAGCACUGACAUAAGUUUGCCACUGAGGCCUUGUGUCAUCGCCACAUUCGCUUUGUCAAUUUGAUCAUCAAUUUUCGCAUGAAAUUUGUACUAC---------AAUAA (((((.(((.....((..(((((((((...(((.....))))))))))))..)).....)))))))).....(((.....(((.((((....)))).))).....)---------))... ( -25.70) >DroWil_CAF1 54426 111 - 1 CAAAGUCGACUCUAGCACUGACAUAAGUUUACCGCUCAGCCCUUGUGUCAUGGCUACAUUCGCCUUGUCGAUUUGAUCGUCGAUUUUUGCGUGAAAUUUGUACUGC---------AUGCA .(((((((((..((((..(((((((((..(........)..)))))))))..))))((.(((......)))..))...)))))))))((((((............)---------))))) ( -29.30) >DroMoj_CAF1 13710 111 - 1 CAAAGUCGACUCCAGCACUGACAUAAGUUUUCCACUUAAACCUUGUGUCAUGGCUACAUUCGCUUUGUCGAUUUGUUCAUCGAUUCUUGCAUGGAAUUUGUACUAA---------AAGAU ...((((((.((((((..(((((((((.(((......))).)))))))))..)))......((...((((((......))))))....))..)))..))).)))..---------..... ( -26.80) >DroPer_CAF1 27325 107 - 1 CAAAGUCGACUCUAGCACUGACAUAAGUUUACCACUUAGCCCUUGUGUCAUCGCUACAUUCGCUUUGUCGAUUUGGUCGUCGAUUUUUGCAUGGAAUUUAUACUAG-------------G (((((.(((.(.((((..(((((((((..(........)..)))))))))..)))).).))))))))(((((......))))).......................-------------. ( -24.40) >consensus CAAAGUCGACUCUAGCACUGACAUAAGUUUACCACUUAGACCUUGUGUCAUCGCUACAUUCGCUUUGUCGAUUUGAUCAUCGAUUUUUGCAUGAAAUUUGUACUAA_________AUGAA (((((.(((.(.((((..(((((((((..(........)..)))))))))..)))).).))))))))(((((......)))))..................................... (-19.44 = -20.33 + 0.89)

| Location | 1,268,400 – 1,268,520 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -38.24 |
| Consensus MFE | -28.19 |
| Energy contribution | -27.72 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1268400 120 + 1281640 AUGACCAAAUCGACAAAGCGAAUGUAGCUAUGACACAAGGUCUAAGCGGUAAACUUAUGUCUGUGCUAGAGUCGACUUUGUCUAAGCUAGCACGAUACGAUGAAGGUAGCUUGAUCGGCU ....((.....((((((((((.(.((((...((((.(((..((....))....))).))))...)))).).))).)))))))(((((((.(.............).)))))))...)).. ( -33.52) >DroVir_CAF1 12040 120 + 1 AUGAUCAAAUCGACAAAGCGAAUGUAGCGAUGACACAAGGAUUAAGUGGCAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCGAAGUUGGCGCGUUACGAUGAAGGUAGCCUCAUUGGCU ..(.((((.((((((((((((.(.(((((.(((((.(((..............))).))))).))))).).))).)))))))))..)))))......((((((.((...))))))))... ( -38.44) >DroGri_CAF1 14161 120 + 1 AUGAUCAAAUUGACAAAGCGAAUGUGGCGAUGACACAAGGCCUCAGUGGCAAACUUAUGUCAGUGCUGGAGUCGACUUUGUCAAAAUUGGCCCGUUACGAUGAAGGUAGCCUAAUUGGCU ...(((...((((((((((((.(.(((((.(((((.((((((.....)))...))).))))).))))).).))).))))))))).((((........))))...)))((((.....)))) ( -39.00) >DroWil_CAF1 54457 120 + 1 ACGAUCAAAUCGACAAGGCGAAUGUAGCCAUGACACAAGGGCUGAGCGGUAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCAAAAUUGGCCCGCUAUGAUGAGGGCAGCCUAAUUGGUU ..((((((...((((((((((.(.((((..(((((.(((.(((....)))...))).)))))..)))).).))).))))))).......((((.(......).)))).......)))))) ( -40.90) >DroMoj_CAF1 13741 120 + 1 AUGAACAAAUCGACAAAGCGAAUGUAGCCAUGACACAAGGUUUAAGUGGAAAACUUAUGUCAGUGCUGGAGUCGACUUUGUCGAAGUUAGGGCGUUACGAUGAGGGCAGUCUCAUUGGAU ...(((...((((((((((((.(.((((..(((((..((((((.......)))))).)))))..)))).).))).))))))))).((....))))).((((((((....))))))))... ( -39.20) >DroPer_CAF1 27352 120 + 1 ACGACCAAAUCGACAAAGCGAAUGUAGCGAUGACACAAGGGCUAAGUGGUAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCAAAAUUAGCUCGAUACGACGAAGGAAGCUUGAUUGGUU ..((((((.((((((((((((.(.(((((.(((((.(((.(((....)))...))).))))).))))).).))).)))))))......((((((...)))(....)..))).)))))))) ( -38.40) >consensus AUGAUCAAAUCGACAAAGCGAAUGUAGCGAUGACACAAGGGCUAAGUGGUAAACUUAUGUCAGUGCUAGAGUCGACUUUGUCAAAAUUAGCCCGUUACGAUGAAGGUAGCCUAAUUGGCU ...........((((((((((.(.((((..(((((.(((..............))).)))))..)))).).))).)))))))...............((((.(((....))).))))... (-28.19 = -27.72 + -0.47)

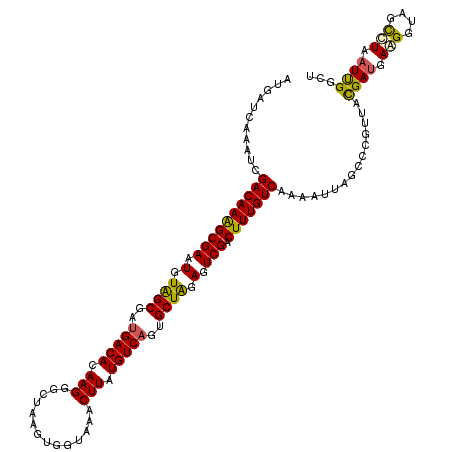
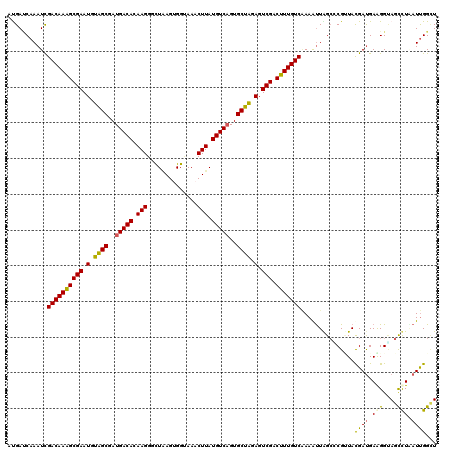
| Location | 1,268,400 – 1,268,520 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -26.81 |
| Energy contribution | -27.03 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
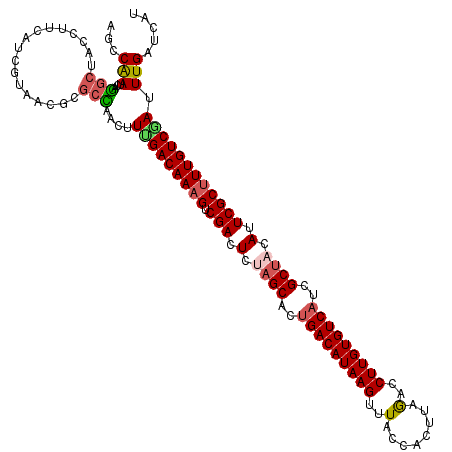
>4_DroMel_CAF1 1268400 120 - 1281640 AGCCGAUCAAGCUACCUUCAUCGUAUCGUGCUAGCUUAGACAAAGUCGACUCUAGCACAGACAUAAGUUUACCGCUUAGACCUUGUGUCAUAGCUACAUUCGCUUUGUCGAUUUGGUCAU .(((((((((((((....(((......))).)))))).(((((((.(((.(.((((...((((((((((((.....)))).))))))))...)))).).)))))))))))).)))))... ( -36.60) >DroVir_CAF1 12040 120 - 1 AGCCAAUGAGGCUACCUUCAUCGUAACGCGCCAACUUCGACAAAGUCGACUCUAGCACUGACAUAAGUUUGCCACUUAAUCCUUGUGUCAUCGCUACAUUCGCUUUGUCGAUUUGAUCAU ((((.....))))..................(((..(((((((((.(((.(.((((..(((((((((..(........)..)))))))))..)))).).)))))))))))).)))..... ( -37.80) >DroGri_CAF1 14161 120 - 1 AGCCAAUUAGGCUACCUUCAUCGUAACGGGCCAAUUUUGACAAAGUCGACUCCAGCACUGACAUAAGUUUGCCACUGAGGCCUUGUGUCAUCGCCACAUUCGCUUUGUCAAUUUGAUCAU ((((.....))))...............(..(((..(((((((((.(((.....((..(((((((((...(((.....))))))))))))..)).....)))))))))))).)))..).. ( -38.20) >DroWil_CAF1 54457 120 - 1 AACCAAUUAGGCUGCCCUCAUCAUAGCGGGCCAAUUUUGACAAAGUCGACUCUAGCACUGACAUAAGUUUACCGCUCAGCCCUUGUGUCAUGGCUACAUUCGCCUUGUCGAUUUGAUCGU ..((.....))..((((.(......).))))(((..(((((((.(.(((.(.((((..(((((((((..(........)..)))))))))..)))).).)))).))))))).)))..... ( -32.50) >DroMoj_CAF1 13741 120 - 1 AUCCAAUGAGACUGCCCUCAUCGUAACGCCCUAACUUCGACAAAGUCGACUCCAGCACUGACAUAAGUUUUCCACUUAAACCUUGUGUCAUGGCUACAUUCGCUUUGUCGAUUUGUUCAU .....(((((......)))))...........(((.(((((((((.(((....(((..(((((((((.(((......))).)))))))))..)))....))))))))))))...)))... ( -34.00) >DroPer_CAF1 27352 120 - 1 AACCAAUCAAGCUUCCUUCGUCGUAUCGAGCUAAUUUUGACAAAGUCGACUCUAGCACUGACAUAAGUUUACCACUUAGCCCUUGUGUCAUCGCUACAUUCGCUUUGUCGAUUUGGUCGU .(((((((.(((((.............)))))......(((((((.(((.(.((((..(((((((((..(........)..)))))))))..)))).).)))))))))))).)))))... ( -34.72) >consensus AGCCAAUCAGGCUACCUUCAUCGUAACGCGCCAACUUUGACAAAGUCGACUCUAGCACUGACAUAAGUUUACCACUUAGACCUUGUGUCAUCGCUACAUUCGCUUUGUCGAUUUGAUCAU ...(((...(((.................)))....(((((((((.(((.(.((((..(((((((((..(........)..)))))))))..)))).).)))))))))))).)))..... (-26.81 = -27.03 + 0.22)

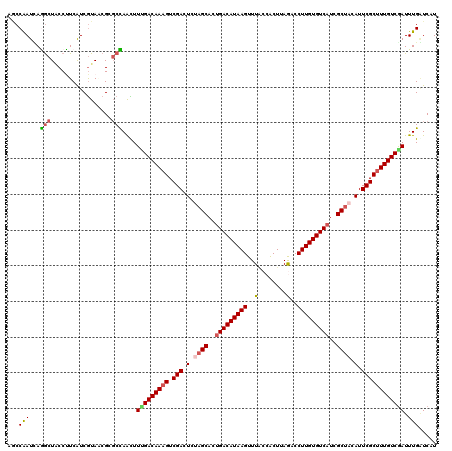
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:02 2006