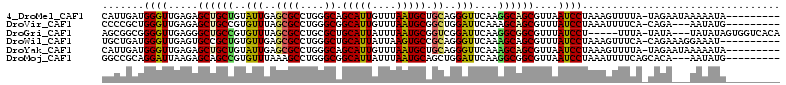
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 1,265,737 – 1,265,840 |
| Length | 103 |
| Max. P | 0.982847 |

| Location | 1,265,737 – 1,265,840 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -15.88 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
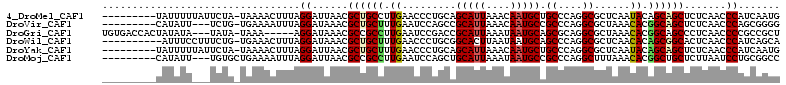
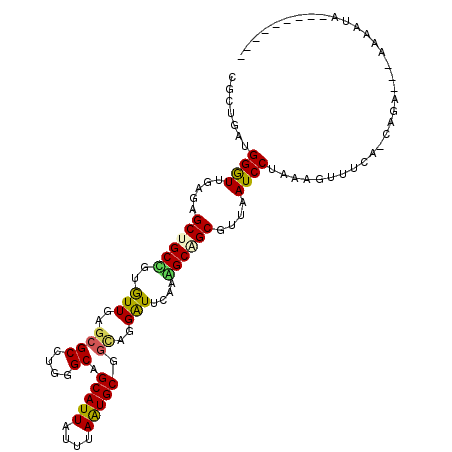
>4_DroMel_CAF1 1265737 103 + 1281640 ---------UAUUUUUAUUCUA-UAAAACUUUAGGAUUAACGCUGCCUUGAACCCUGCAGCAUUAAACAAUGCUGCCCAGGCGCUCAAUACAGCAGCUCUCAACCCAUCAAUG ---------.......((((((-........))))))....(((((.((((.((((((((((((....))))))))..))).).))))....)))))................ ( -26.80) >DroVir_CAF1 9078 100 + 1 ---------CAUAUU---UCUG-UGAAAAUUUAGGAUAAACGCUGCUUUGAAUCCAGCCGCAUUAAACAAUGCCGCCCAGGCGCUAAACACGGCAGCUCUCAACCCAGCGGGG ---------.....(---((((-........)))))....(((((..((((.....((((((((....)))))(((....)))........))).....))))..)))))... ( -25.30) >DroGri_CAF1 12665 104 + 1 UGUGACCACUAUAUA---UAUA-UAAA-----AGGAUAAACGCCGCCUUGAAUCCGACCGCAUUAAAUAAUGCAGCGCAGGCGCUAAACACGGCAGCCCUCAACCCCGCCGCU .(((.((..((((..---..))-))..-----.))......((.(((.((.........(((((....)))))((((....))))...)).))).))............))). ( -20.60) >DroWil_CAF1 53219 102 + 1 ----------AUUUCCUUUCUG-UGAAACUUUAGGAUAAACGCUGCUUUGAACCCUGCGGCACUUAAUAAUGCAGCCCAGGCGCUCAACACAGCGGCACUCAACCCAUCAGCA ----------...((((....(-.....)...))))....(((((..((((.(((((.(((.............))))))).).))))..))))).................. ( -24.82) >DroYak_CAF1 17594 103 + 1 ---------UAUUUUUAUUCUA-UAAAACUUUAGGAUUAACGCUGCUUUGAACCCUGCAGCAUUAAACAAUGCUGCCCAGGCGCUCAAUACAGCAGCUCUCAACCCAUCAAUG ---------.......((((((-........))))))....((((((((((.((((((((((((....))))))))..))).).))))...))))))................ ( -28.70) >DroMoj_CAF1 11580 101 + 1 ---------CAUAUU---UGUGCUGAAAAUUUAGGAUUAACGCCGCCUUGAAUCCAGCUGCAUUAAAUAAUGCCGCCCAGGCUUUAAACACGGCUGCUCUUAAUCCUGCGGCC ---------......---...((((......(((((((((.((.((((((((.((.((.(((((....))))).))...)).)))))....))).))..))))))))))))). ( -27.70) >consensus _________UAUAUU___UCUA_UAAAACUUUAGGAUAAACGCUGCCUUGAACCCUGCAGCAUUAAACAAUGCAGCCCAGGCGCUAAACACAGCAGCUCUCAACCCAGCAGCG .................................((......((((((.((.........(((((....))))).((....))......)).)))))).......))....... (-15.88 = -15.90 + 0.03)
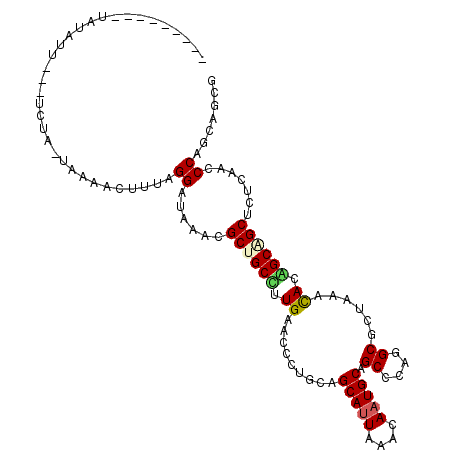
| Location | 1,265,737 – 1,265,840 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -20.38 |
| Energy contribution | -19.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1265737 103 - 1281640 CAUUGAUGGGUUGAGAGCUGCUGUAUUGAGCGCCUGGGCAGCAUUGUUUAAUGCUGCAGGGUUCAAGGCAGCGUUAAUCCUAAAGUUUUA-UAGAAUAAAAAUA--------- .......(((((((..((((((...((((((.(((..((((((((....))))))))))))))))))))))).)))))))..........-.............--------- ( -37.70) >DroVir_CAF1 9078 100 - 1 CCCCGCUGGGUUGAGAGCUGCCGUGUUUAGCGCCUGGGCGGCAUUGUUUAAUGCGGCUGGAUUCAAAGCAGCGUUUAUCCUAAAUUUUCA-CAGA---AAUAUG--------- ......((((.((((.(((((..((((((((((....)).(((((....))))).))))))..))..))))).)))).))))........-....---......--------- ( -28.60) >DroGri_CAF1 12665 104 - 1 AGCGGCGGGGUUGAGGGCUGCCGUGUUUAGCGCCUGCGCUGCAUUAUUUAAUGCGGUCGGAUUCAAGGCGGCGUUUAUCCU-----UUUA-UAUA---UAUAUAGUGGUCACA .(.(((((((.((((.(((((((((.....)))(((.((((((((....)))))))))))......)))))).)))).)))-----)(((-((..---..)))))..))).). ( -34.90) >DroWil_CAF1 53219 102 - 1 UGCUGAUGGGUUGAGUGCCGCUGUGUUGAGCGCCUGGGCUGCAUUAUUAAGUGCCGCAGGGUUCAAAGCAGCGUUUAUCCUAAAGUUUCA-CAGAAAGGAAAU---------- .(((..((((.((((...((((((.((((((.(((((((.((........))))).)))))))))).)))))))))).)))).)))....-............---------- ( -35.00) >DroYak_CAF1 17594 103 - 1 CAUUGAUGGGUUGAGAGCUGCUGUAUUGAGCGCCUGGGCAGCAUUGUUUAAUGCUGCAGGGUUCAAAGCAGCGUUAAUCCUAAAGUUUUA-UAGAAUAAAAAUA--------- .......(((((((..((((((...((((((.(((..((((((((....))))))))))))))))))))))).)))))))..........-.............--------- ( -37.70) >DroMoj_CAF1 11580 101 - 1 GGCCGCAGGAUUAAGAGCAGCCGUGUUUAAAGCCUGGGCGGCAUUAUUUAAUGCAGCUGGAUUCAAGGCGGCGUUAAUCCUAAAUUUUCAGCACA---AAUAUG--------- ......((((((((.....(((((.((.....((..(((.(((((....))))).)))))....)).))))).))))))))..............---......--------- ( -26.80) >consensus CGCUGAUGGGUUGAGAGCUGCCGUGUUGAGCGCCUGGGCAGCAUUAUUUAAUGCGGCAGGAUUCAAAGCAGCGUUAAUCCUAAAGUUUCA_CAGA___AAAAUA_________ .......((((.....((((((..(((..((((....)).(((((....))))).))..)))....))))))....))))................................. (-20.38 = -19.93 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:58 2006