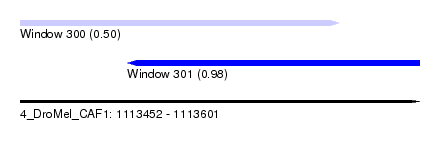
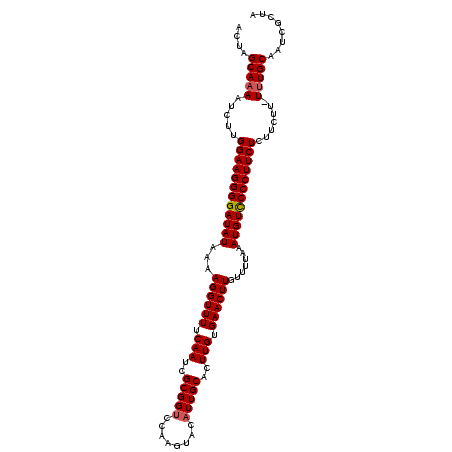
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 1,113,452 – 1,113,601 |
| Length | 149 |
| Max. P | 0.976391 |

| Location | 1,113,452 – 1,113,571 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -23.82 |
| Energy contribution | -23.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1113452 119 + 1281640 AUGGUAAUUCCAGUUGCAACCUUCCAAAACUAAAGAACAAUAGCAAUUGCAGA-AAGAAGAGAAGGGAACAUUUUAAACAAGUUCACAAGUGCAAUGCACUUGGGCCGCGAUUGAAAACC ..(((..((.(((((((..((((((....(....).......((....))...-.....).)))))((((...........)))).(((((((...)))))))....))))))).))))) ( -26.20) >DroSec_CAF1 26398 119 + 1 AUGGUAAUUCCAGUUGCAACCGUCCAAAACUUAAGAACAAUAGCGAUUGCAAA-AAGAAGAGAAGGGGACAUUUAAAACAAGUUCACAAGUGCAAUGUACUUGGACCGCGAUUGAAAACC ..(((..((.(((((((....((((....(((..........((....))...-........))).))))...........(.((.(((((((...))))))))).)))))))).))))) ( -24.20) >DroSim_CAF1 14032 120 + 1 AUGGUAAUUCCAGUUGCAAGCGUCCAAAACUUAAGAACAAUAGCGAUUGCAAAAAAGAAGAGAAGGGGACAUUUAAAACAAGUUCACAAGUGCAAUGUACUUGGACCGCGAUUGAAAACC ..(((..((.(((((((..(.((((.........((((....((....))..............(....)...........))))..((((((...)))))))))))))))))).))))) ( -24.10) >consensus AUGGUAAUUCCAGUUGCAACCGUCCAAAACUUAAGAACAAUAGCGAUUGCAAA_AAGAAGAGAAGGGGACAUUUAAAACAAGUUCACAAGUGCAAUGUACUUGGACCGCGAUUGAAAACC ..(((..((.(((((((.................((((....((....))..............(....)...........)))).(((((((...)))))))....))))))).))))) (-23.82 = -23.60 + -0.22)

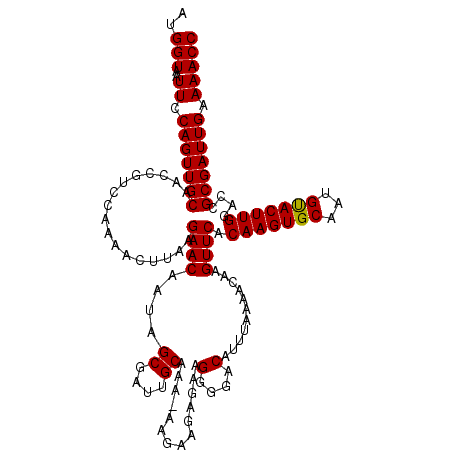
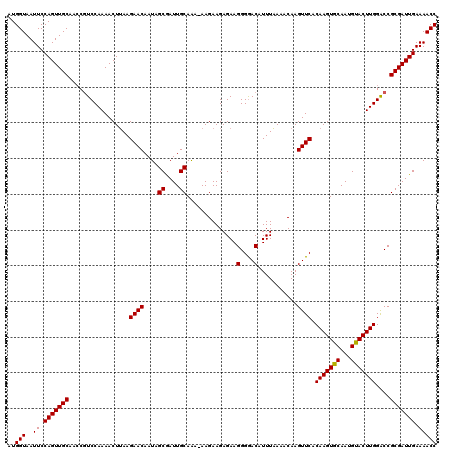
| Location | 1,113,492 – 1,113,601 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -28.65 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1113492 109 - 1281640 ACUAGCAAAAUCUUGGAAGGGGAUAUAAAAGGUUUUCAAUCGCGGCCCAAGUGCAUUGCACUUGUGAACUUGUUUAAAAUGUUCCCUUCUCUUCUU-UCUGCAAUUGCUA ....(((.((....((((((((((((....((((.........))))(((((((...)))))))..............)))))))))))).....)-).)))........ ( -28.30) >DroSec_CAF1 26438 109 - 1 ACCAGCAAAAUCUUGGAAGGGGAUAUAAAAGGUUUUCAAUCGCGGUCCAAGUACAUUGCACUUGUGAACUUGUUUUAAAUGUCCCCUUCUCUUCUU-UUUGCAAUCGCUA ....((((((....((((((((((((...((((((.(((..(((((........)))))..))).)))))).......)))))))))))).....)-)))))........ ( -33.10) >DroSim_CAF1 14072 110 - 1 ACUAGCAAAAUCUUGGAAGGGGAUAUAAAAGGUUUUCAAUCGCGGUCCAAGUACAUUGCACUUGUGAACUUGUUUUAAAUGUCCCCUUCUCUUCUUUUUUGCAAUCGCUA ....((((((....((((((((((((...((((((.(((..(((((........)))))..))).)))))).......))))))))))))......))))))........ ( -32.50) >consensus ACUAGCAAAAUCUUGGAAGGGGAUAUAAAAGGUUUUCAAUCGCGGUCCAAGUACAUUGCACUUGUGAACUUGUUUUAAAUGUCCCCUUCUCUUCUU_UUUGCAAUCGCUA ....(((((.....((((((((((((...((((((.(((..(((((........)))))..))).)))))).......)))))))))))).......)))))........ (-28.65 = -29.10 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:38 2006