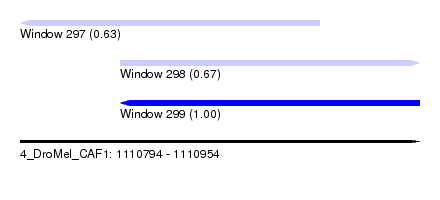
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 1,110,794 – 1,110,954 |
| Length | 160 |
| Max. P | 0.998281 |

| Location | 1,110,794 – 1,110,914 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -23.62 |
| Energy contribution | -23.62 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
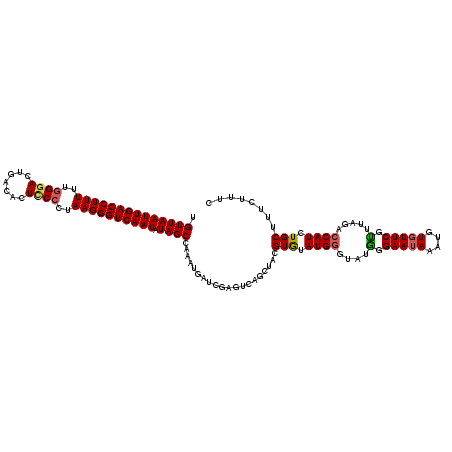
>4_DroMel_CAF1 1110794 120 - 1281640 UGUUAAUUGACCUUUUUGGGACUGACACUCCCCUAAGGGUCAAUAUAGCUAAAUGAUCGAGUCAGCUACGUGUAUGGGUAUGGGAAUAAAUGUGUUCGUUUAGGCCAUCUGCUUUCUUUC .((((((((((((((..((((.......))))..)))))))))).)))).........(((..(((..((((......))))((..((((((....))))))..))....)))..))).. ( -32.60) >DroSec_CAF1 23871 120 - 1 UGUUAAUUGACCUUUUUGGGACUGACACUUCCUUAAGGGUCAAUAUAGCCAUAUGAUCGAGUCAGCUACGUGUAUGGGUAUAGGAAUAAAUGUGUUCGUUUAGCCCAUCUGCUUUCUUUC ......(((((((((..((((.......))))..)))))))))((((.((((((...((((....)).)).)))))).))))((..((((((....))))))..)).............. ( -32.90) >DroSim_CAF1 11523 119 - 1 UGUUAAUUGACCUUUUUGGGACUGACACUUC-UUAAGGGUCAAUAUAGCCAUAUGAUCGAGUCAGCUACGUGUAUGGGUAUAGGAAUAAAUGUGUUCGUUUAGCCCAUCUGCUUUCUUUC ......(((((((((..(((........)))-..)))))))))((((.((((((...((((....)).)).)))))).))))((..((((((....))))))..)).............. ( -29.40) >DroEre_CAF1 24101 120 - 1 UGUUAAUUGACCUUUUUGGGACCAUCACUCCCCUAAGGGUCAAUAUAGCUAAAUGAACGAGUCAGCUACGUACAUGAGUAUGGGAAUAAAUGUGUUCGCUUAGACCAUCUGCUUUAUUUU .((((((((((((((..((((.......))))..)))))))))).)))).((((((((((((((((..(((((....))))).(((((....))))))))..)))..)).).))))))). ( -32.20) >DroYak_CAF1 15371 116 - 1 UGUUAAUUGACCUUUUUGGAACCAUCACUCCCCUAAGGGUCAAUAUAGCAAAAUGAACGA----GCUACGUACAUGGGUAUGAGAAGAAAUGUGUUCGUUUAGACCAUCUGCUUUAUUUU (((((((((((((((..(((........)))...)))))))))).)))))...(((((((----((..(((((....)))))...(....)..))))))))).................. ( -30.00) >consensus UGUUAAUUGACCUUUUUGGGACUGACACUCCCCUAAGGGUCAAUAUAGCCAAAUGAUCGAGUCAGCUACGUGUAUGGGUAUGGGAAUAAAUGUGUUCGUUUAGACCAUCUGCUUUCUUUC .((((((((((((((..((((.......))))..)))))))))).))))....................(((.((((....(.(((((....))))).).....)))).)))........ (-23.62 = -23.62 + -0.00)

| Location | 1,110,834 – 1,110,954 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -25.90 |
| Energy contribution | -26.26 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1110834 120 + 1281640 UACCCAUACACGUAGCUGACUCGAUCAUUUAGCUAUAUUGACCCUUAGGGGAGUGUCAGUCCCAAAAAGGUCAAUUAACAACAGGUGAAUAAGUGCUAGCAACCACGUUUGCGCGGGUUA .((((...(((((((((((.........))))))))(((((((.((..((((.......))))..)).))))))).........))).....((((.(((......))).)))))))).. ( -37.10) >DroSec_CAF1 23911 120 + 1 UACCCAUACACGUAGCUGACUCGAUCAUAUGGCUAUAUUGACCCUUAAGGAAGUGUCAGUCCCAAAAAGGUCAAUUAACAACAGGUGAAAAAGUGCUAGGGCGCACGUUUGCGCGGGUUA .((((.....(.((((..(((...((((.((.....(((((((.....(((........)))......)))))))......)).))))...))))))).)(((((....))))))))).. ( -34.40) >DroSim_CAF1 11563 119 + 1 UACCCAUACACGUAGCUGACUCGAUCAUAUGGCUAUAUUGACCCUUAA-GAAGUGUCAGUCCCAAAAAGGUCAAUUAACAACAGGUGAAAAAGUGCUAGCACGCACGUUUGCGCGCGUUA .........((((.((.(((....((((.((.....(((((((.....-...................)))))))......)).))))....((((......))))))).))..)))).. ( -24.36) >DroEre_CAF1 24141 120 + 1 UACUCAUGUACGUAGCUGACUCGUUCAUUUAGCUAUAUUGACCCUUAGGGGAGUGAUGGUCCCAAAAAGGUCAAUUAACAACAGGUGAAUGAGUGCUAGCAAACACGUUUGCGCGGGUUC .((((.......((((..((((((((((((......(((((((.((..((((.......))))..)).))))))).......))))))))))))))))(((((....)))))..)))).. ( -39.62) >DroYak_CAF1 15411 116 + 1 UACCCAUGUACGUAGC----UCGUUCAUUUUGCUAUAUUGACCCUUAGGGGAGUGAUGGUUCCAAAAAGGUCAAUUAACAACAGGUGAAUGAGUGCUAGCAAACACGUUUGCGCGGGUUC .((((.........((----((((((((((......(((((((......((((......)))).....))))))).......))))))))))))((..(((((....))))))))))).. ( -37.02) >consensus UACCCAUACACGUAGCUGACUCGAUCAUUUAGCUAUAUUGACCCUUAGGGGAGUGUCAGUCCCAAAAAGGUCAAUUAACAACAGGUGAAUAAGUGCUAGCAAACACGUUUGCGCGGGUUA .((((...(((((((((.............))))))(((((((.((..((((.(...).))))..)).))))))).........))).....((((.(((......))).)))))))).. (-25.90 = -26.26 + 0.36)

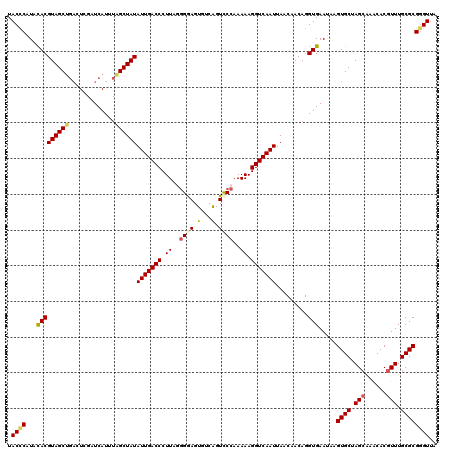
| Location | 1,110,834 – 1,110,954 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.94 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
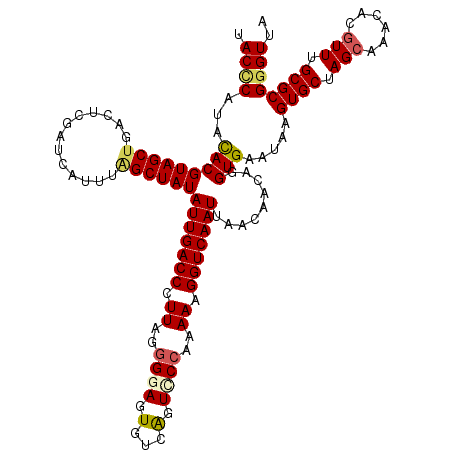
>4_DroMel_CAF1 1110834 120 - 1281640 UAACCCGCGCAAACGUGGUUGCUAGCACUUAUUCACCUGUUGUUAAUUGACCUUUUUGGGACUGACACUCCCCUAAGGGUCAAUAUAGCUAAAUGAUCGAGUCAGCUACGUGUAUGGGUA ..(((((..(..(((((((((.....((((..(((...(((((..((((((((((..((((.......))))..)))))))))))))))....)))..))))))))))))))..))))). ( -44.40) >DroSec_CAF1 23911 120 - 1 UAACCCGCGCAAACGUGCGCCCUAGCACUUUUUCACCUGUUGUUAAUUGACCUUUUUGGGACUGACACUUCCUUAAGGGUCAAUAUAGCCAUAUGAUCGAGUCAGCUACGUGUAUGGGUA ..(((((((((....)))))..((((((((..(((..(((.((((((((((((((..((((.......))))..)))))))))).)))).))))))..))))..)))).......)))). ( -40.20) >DroSim_CAF1 11563 119 - 1 UAACGCGCGCAAACGUGCGUGCUAGCACUUUUUCACCUGUUGUUAAUUGACCUUUUUGGGACUGACACUUC-UUAAGGGUCAAUAUAGCCAUAUGAUCGAGUCAGCUACGUGUAUGGGUA ..(((((((((....)))))))((((((((..(((..(((.((((((((((((((..(((........)))-..)))))))))).)))).))))))..))))..)))).))......... ( -34.70) >DroEre_CAF1 24141 120 - 1 GAACCCGCGCAAACGUGUUUGCUAGCACUCAUUCACCUGUUGUUAAUUGACCUUUUUGGGACCAUCACUCCCCUAAGGGUCAAUAUAGCUAAAUGAACGAGUCAGCUACGUACAUGAGUA .............(((((.((.((((((((.((((...(((((..((((((((((..((((.......))))..)))))))))))))))....)))).))))..)))))).))))).... ( -38.00) >DroYak_CAF1 15411 116 - 1 GAACCCGCGCAAACGUGUUUGCUAGCACUCAUUCACCUGUUGUUAAUUGACCUUUUUGGAACCAUCACUCCCCUAAGGGUCAAUAUAGCAAAAUGAACGA----GCUACGUACAUGGGUA ..(((((((((((....)))))..)).(((.((((....((((((((((((((((..(((........)))...)))))))))).))))))..)))).))----)..........)))). ( -37.00) >consensus UAACCCGCGCAAACGUGCUUGCUAGCACUUAUUCACCUGUUGUUAAUUGACCUUUUUGGGACUGACACUCCCCUAAGGGUCAAUAUAGCCAAAUGAUCGAGUCAGCUACGUGUAUGGGUA ..((((((((....)))).(((((((((((..(((...(((((..((((((((((..((((.......))))..)))))))))))))))....)))..))))..)))).)))...)))). (-31.30 = -31.94 + 0.64)

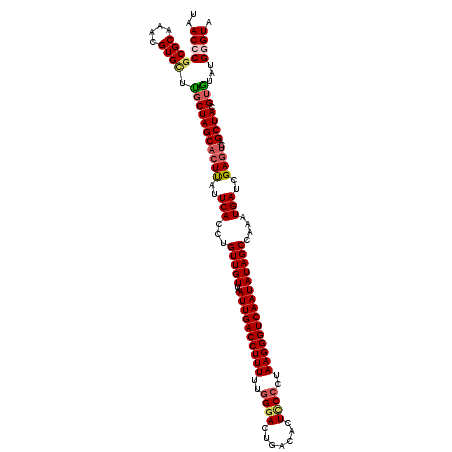
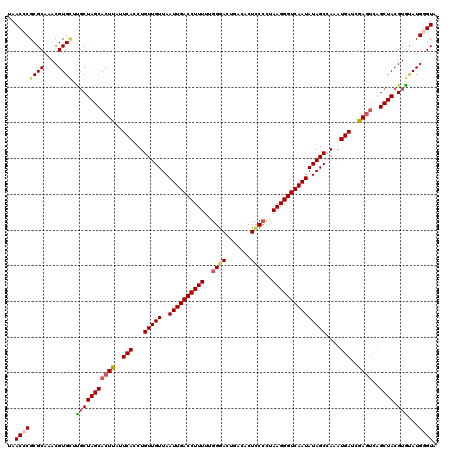
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:36 2006