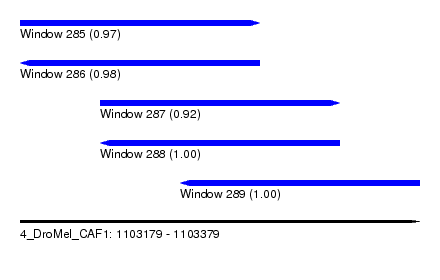
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 1,103,179 – 1,103,379 |
| Length | 200 |
| Max. P | 0.999385 |

| Location | 1,103,179 – 1,103,299 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -39.66 |
| Consensus MFE | -32.02 |
| Energy contribution | -32.14 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
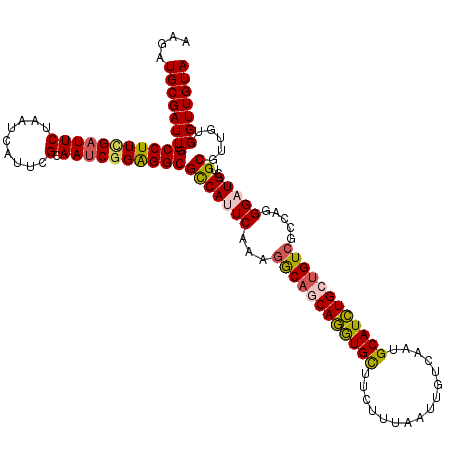
>4_DroMel_CAF1 1103179 120 + 1281640 CGCAUUGCAGCGGGUUGUGGAAAUGACUUCCGGGAGCCAAGGCUGUUGCAGCAGUUUAAGUUAAAGCAGCAACAGCACUUUACAACCACAACGCAGCAUCCUUGGCGACAGCAGAUGCAU .(((((((..(((((..(....)..))..)))(..(((((((.((((((....((((......)))).((....))................)))))).)))))))..).))).)))).. ( -42.60) >DroSec_CAF1 16940 120 + 1 CGCAUUGCAGCGGAUUGUGGAAACGACUUCCGGGAGCCAAGGCUGUUGCAGCAGUUUAAGUUAAAGCAGCAACAGCACUUUACAACCACAACGCAGCAUCCCUGGCGACAGCAGAUGCAU .((.(((....(((...((....))...)))((..(..(((((((((((.((.............)).)))))))).)))..)..)).))).)).(((((.(((....)))..))))).. ( -41.52) >DroSim_CAF1 7573 120 + 1 CGCAUUGCAGCAGAUUGUGGAAACGACUUCCGGGAGCCAAGGUUGUUGCAGCAGUUUAAGUUAAAGCAGCAACAGCACUUUACAACCACAACGCAGCAUCCCUGGCGACAGCAGAUGCAU .(((((((......((.(((((.....))))).))((((.((.((((((....((((......)))).((....))................)))))).)).))))....))).)))).. ( -35.90) >DroEre_CAF1 16494 120 + 1 CGCAUUACAGCAGGCUGUGAAAAUAACUUCCGUGAGCCAAGGCUGUUGCAGCAGUUAAAGCUAUAGCAGCAGCAGCACUUUACAACCACAACGCAGCAGCCCUGGCGACAGCAGAUGCAU .........(((.((((((((......))).....((((.((((((((((((.......)))...((....))...................))))))))).)))).)))))...))).. ( -40.50) >DroYak_CAF1 8476 120 + 1 CGCAUUACAGCAGGCUGUGGAAAUGACUUUCGGGAUCCCAGGCUGUUGCAACAGUUUAAGCUAUAGCAGCAGCAGCACUUUACAACCACAACGCAGCAUCCCUGGCGACAGCAAAUGCAU .(((((.(((...((((((((((....))))((....))..((((((((...(((....)))...))))))))..................))))))....)))((....)).))))).. ( -37.80) >consensus CGCAUUGCAGCAGGUUGUGGAAAUGACUUCCGGGAGCCAAGGCUGUUGCAGCAGUUUAAGUUAAAGCAGCAACAGCACUUUACAACCACAACGCAGCAUCCCUGGCGACAGCAGAUGCAU .(((((.(((...(((((((((.....))))((.....(((((((((((.((.............)).)))))))).))).....)).....)))))....)))((....)).))))).. (-32.02 = -32.14 + 0.12)

| Location | 1,103,179 – 1,103,299 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -42.45 |
| Consensus MFE | -35.96 |
| Energy contribution | -36.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1103179 120 - 1281640 AUGCAUCUGCUGUCGCCAAGGAUGCUGCGUUGUGGUUGUAAAGUGCUGUUGCUGCUUUAACUUAAACUGCUGCAACAGCCUUGGCUCCCGGAAGUCAUUUCCACAACCCGCUGCAAUGCG ..((((((((....))...)))))).(((((((((((((.(((.((((((((.((.............)).))))))))))).......(((((...)))))))))))....))))))). ( -44.12) >DroSec_CAF1 16940 120 - 1 AUGCAUCUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUAAAGUGCUGUUGCUGCUUUAACUUAAACUGCUGCAACAGCCUUGGCUCCCGGAAGUCGUUUCCACAAUCCGCUGCAAUGCG ..((((((.(((....))))))))).((((((..((.((.(((.((((((((.((.............)).))))))))))).))....(((((...))))).......))..)))))). ( -44.62) >DroSim_CAF1 7573 120 - 1 AUGCAUCUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUAAAGUGCUGUUGCUGCUUUAACUUAAACUGCUGCAACAACCUUGGCUCCCGGAAGUCGUUUCCACAAUCUGCUGCAAUGCG ..((((.(((.((.(((((((.((.((((..(..(((.((((((((....)).)))))).....)))..))))).)).)))))))....(((((...))))).......)).))))))). ( -40.40) >DroEre_CAF1 16494 120 - 1 AUGCAUCUGCUGUCGCCAGGGCUGCUGCGUUGUGGUUGUAAAGUGCUGCUGCUGCUAUAGCUUUAACUGCUGCAACAGCCUUGGCUCACGGAAGUUAUUUUCACAGCCUGCUGUAAUGCG .((((.(.(((((.((((((((((.((((..(..((..((((((...((....))....))))))))..))))).))))))))))..((....)).......)))))..).))))..... ( -45.70) >DroYak_CAF1 8476 120 - 1 AUGCAUUUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUAAAGUGCUGCUGCUGCUAUAGCUUAAACUGUUGCAACAGCCUGGGAUCCCGAAAGUCAUUUCCACAGCCUGCUGUAAUGCG ..((((((.(((....))))))))).((((((..((.....((.(((((((.(((.((((......)))).))).)))).(((((...(....)....))))).)))))))..)))))). ( -37.40) >consensus AUGCAUCUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUAAAGUGCUGUUGCUGCUUUAACUUAAACUGCUGCAACAGCCUUGGCUCCCGGAAGUCAUUUCCACAACCUGCUGCAAUGCG ..((((((.(((....))))))))).(((((((((((((.(((.((((((((.((.............)).))))))))))).......(((((...)))))))))))....))))))). (-35.96 = -36.20 + 0.24)


| Location | 1,103,219 – 1,103,339 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -34.51 |
| Consensus MFE | -29.36 |
| Energy contribution | -29.48 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1103219 120 + 1281640 GGCUGUUGCAGCAGUUUAAGUUAAAGCAGCAACAGCACUUUACAACCACAACGCAGCAUCCUUGGCGACAGCAGAUGCAUUGACAAUUAAAGAAACACCUGCUGCCUUUGAAUGGCGCCC .((((((((.((.............)).))))))))...............(((.(((((.(((....)))..))))).....((.((((((.............)))))).)))))... ( -30.74) >DroSec_CAF1 16980 120 + 1 GGCUGUUGCAGCAGUUUAAGUUAAAGCAGCAACAGCACUUUACAACCACAACGCAGCAUCCCUGGCGACAGCAGAUGCAUUGACAAUUAAAGAAGCAUCUGCUGCCUUUGAAUGACGCCU .((((((((.((.............)).))))))))................((.(((((.(((....)))..))))).....((.((((((.(((....)))..)))))).))..)).. ( -36.52) >DroSim_CAF1 7613 120 + 1 GGUUGUUGCAGCAGUUUAAGUUAAAGCAGCAACAGCACUUUACAACCACAACGCAGCAUCCCUGGCGACAGCAGAUGCAUUGACAAUUAAAGAAGCAUCUGCUGCCUUUGAAUGACGCCU ((.((((((....((((......)))).((....))................)))))).))..((((.((((((((((.((..........)).)))))))))).(.......).)))). ( -34.40) >DroEre_CAF1 16534 120 + 1 GGCUGUUGCAGCAGUUAAAGCUAUAGCAGCAGCAGCACUUUACAACCACAACGCAGCAGCCCUGGCGACAGCAGAUGCAUUGGCAAUUAAAGAAGCACCUGUUGUUUUUGAAUGGCGCCU ((((((((((((.......)))...((....))...................)))))))))..(((((((((((.(((.((..........)).))).)))))))..........)))). ( -35.90) >DroYak_CAF1 8516 120 + 1 GGCUGUUGCAACAGUUUAAGCUAUAGCAGCAGCAGCACUUUACAACCACAACGCAGCAUCCCUGGCGACAGCAAAUGCAUUGGCAAUUAAAGAAGCACUUGUAGCCUUUGAUUGGCGCCU .((((((((...(((....)))...))))))))...................((.((((..(((....)))...))))...(.(((((((((..((.......))))))))))).))).. ( -35.00) >consensus GGCUGUUGCAGCAGUUUAAGUUAAAGCAGCAACAGCACUUUACAACCACAACGCAGCAUCCCUGGCGACAGCAGAUGCAUUGACAAUUAAAGAAGCACCUGCUGCCUUUGAAUGGCGCCU .((((((((.((.............)).))))))))................((.(((((.(((....)))..)))))...(.((.((((((.(((....)))..)))))).)).))).. (-29.36 = -29.48 + 0.12)

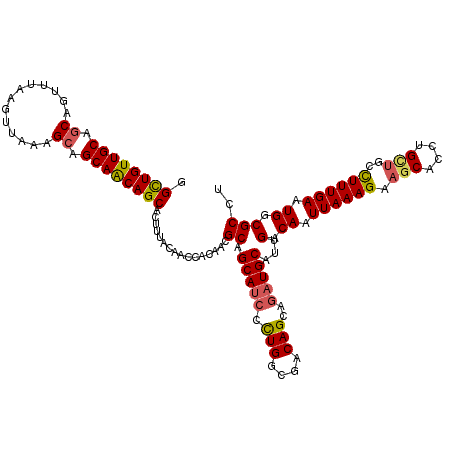
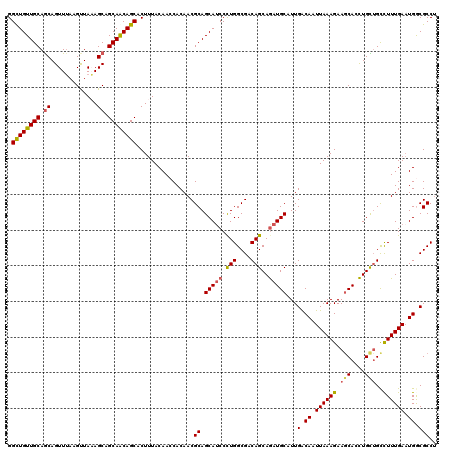
| Location | 1,103,219 – 1,103,339 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -37.29 |
| Energy contribution | -36.97 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.999385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1103219 120 - 1281640 GGGCGCCAUUCAAAGGCAGCAGGUGUUUCUUUAAUUGUCAAUGCAUCUGCUGUCGCCAAGGAUGCUGCGUUGUGGUUGUAAAGUGCUGUUGCUGCUUUAACUUAAACUGCUGCAACAGCC .((((((((((...((((((((((((................)))))))))))).....)))))..))))).............((((((((.((.............)).)))))))). ( -42.71) >DroSec_CAF1 16980 120 - 1 AGGCGUCAUUCAAAGGCAGCAGAUGCUUCUUUAAUUGUCAAUGCAUCUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUAAAGUGCUGUUGCUGCUUUAACUUAAACUGCUGCAACAGCC ..(((.(((((...((((((((((((................)))))))))))).....))))).)))................((((((((.((.............)).)))))))). ( -45.31) >DroSim_CAF1 7613 120 - 1 AGGCGUCAUUCAAAGGCAGCAGAUGCUUCUUUAAUUGUCAAUGCAUCUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUAAAGUGCUGUUGCUGCUUUAACUUAAACUGCUGCAACAACC ..(((.(((((...((((((((((((................)))))))))))).....))))).)))((((..((..((((((((....)).)))))).........))..)))).... ( -44.09) >DroEre_CAF1 16534 120 - 1 AGGCGCCAUUCAAAAACAACAGGUGCUUCUUUAAUUGCCAAUGCAUCUGCUGUCGCCAGGGCUGCUGCGUUGUGGUUGUAAAGUGCUGCUGCUGCUAUAGCUUUAACUGCUGCAACAGCC .(((((....(((..(((((((((((................))))))((.((.((....)).)).)))))))..)))....)))))((((.(((..(((......)))..))).)))). ( -39.09) >DroYak_CAF1 8516 120 - 1 AGGCGCCAAUCAAAGGCUACAAGUGCUUCUUUAAUUGCCAAUGCAUUUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUAAAGUGCUGCUGCUGCUAUAGCUUAAACUGUUGCAACAGCC ....(((.......)))....((..(((...((((..(.((((((.((.(((....))).))...)))))))..))))..)))..))((((.(((.((((......)))).))).)))). ( -36.40) >consensus AGGCGCCAUUCAAAGGCAGCAGGUGCUUCUUUAAUUGUCAAUGCAUCUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUAAAGUGCUGUUGCUGCUUUAACUUAAACUGCUGCAACAGCC ..(((.(((((...((((((((((((................)))))))))))).....))))).)))((((..(..((.((((...((....))....))))..))..)..)))).... (-37.29 = -36.97 + -0.32)

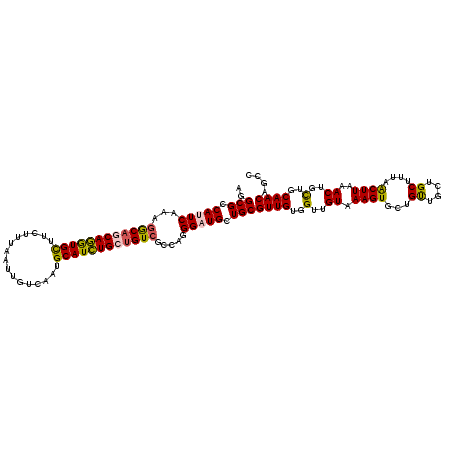
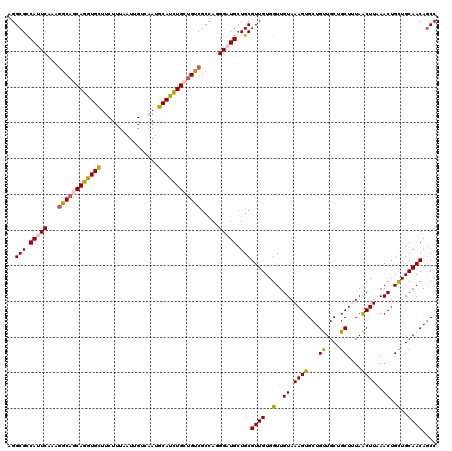
| Location | 1,103,259 – 1,103,379 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -38.64 |
| Consensus MFE | -35.75 |
| Energy contribution | -35.95 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1103259 120 - 1281640 AAGAUGCGAUUUGCCUUCGAUUCUAAUCAUUCGCAAUCGGGGGCGCCAUUCAAAGGCAGCAGGUGUUUCUUUAAUUGUCAAUGCAUCUGCUGUCGCCAAGGAUGCUGCGUUGUGGUUGUA ..((((((....(((((((((((.........).))))))))))((.((((...((((((((((((................)))))))))))).....))))))))))))......... ( -40.09) >DroSec_CAF1 17020 120 - 1 AAGAUGCGAUUUGCCUUCGAUUCUAAUCAUUCGCAAUCGGAGGCGUCAUUCAAAGGCAGCAGAUGCUUCUUUAAUUGUCAAUGCAUCUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUA ..((((((....(((((((((((.........).))))))))))(.(((((...((((((((((((................)))))))))))).....))))))))))))......... ( -44.69) >DroSim_CAF1 7653 120 - 1 AAGAUGCGAUUUGCCUUCGAUUCUAAUCAUUCGCAAUCGGAGGCGUCAUUCAAAGGCAGCAGAUGCUUCUUUAAUUGUCAAUGCAUCUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUA ..((((((....(((((((((((.........).))))))))))(.(((((...((((((((((((................)))))))))))).....))))))))))))......... ( -44.69) >DroEre_CAF1 16574 119 - 1 -AUAUGCGAUUUGCCUUUGAUUCUAAUCAUUCGCAACCGGAGGCGCCAUUCAAAAACAACAGGUGCUUCUUUAAUUGCCAAUGCAUCUGCUGUCGCCAGGGCUGCUGCGUUGUGGUUGUA -................((((....))))...(((((((((((((((..............)))))))))........(((((((...((.(((.....))).))))))))).)))))). ( -33.04) >DroYak_CAF1 8556 119 - 1 -AUAUGCGAUUUGCCUUUGAUUCUAAUUAUUCGCAAUCCGAGGCGCCAAUCAAAGGCUACAAGUGCUUCUUUAAUUGCCAAUGCAUUUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUA -....(((....((((((((((.......((((.....)))).....))))))))))......))).....((((..(.((((((.((.(((....))).))...)))))))..)))).. ( -30.70) >consensus AAGAUGCGAUUUGCCUUCGAUUCUAAUCAUUCGCAAUCGGAGGCGCCAUUCAAAGGCAGCAGGUGCUUCUUUAAUUGUCAAUGCAUCUGCUGUCGCCAGGGAUGCUGCGUUGUGGUUGUA ....(((((((.(((((((((((.........).))))))))))(((((((...((((((((((((................)))))))))))).....)))))..)).....))))))) (-35.75 = -35.95 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:23 2006