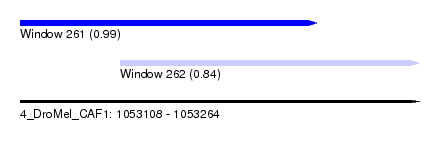
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 1,053,108 – 1,053,264 |
| Length | 156 |
| Max. P | 0.988106 |

| Location | 1,053,108 – 1,053,224 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.82 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -13.47 |
| Energy contribution | -13.28 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
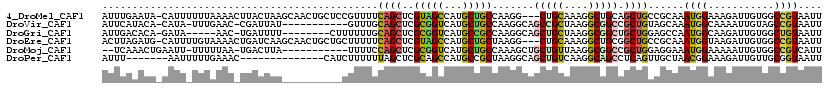
>4_DroMel_CAF1 1053108 116 + 1281640 AUUUGAAUA-CAUUUUUUAAAACUUACUAAGCAACUGCUCCGUUUUCAGCUCGUAGCCAUGCUGCCAAGG---CUGCAAAGGCUGCAGCUGCCGCAAAUGGAAAGAUUGUGGCCGUAAUU .((((((..-.....))))))..((((...((((((..(((((((.(((((.((((((.(((.((....)---).)))..)))))))))))....))))))).)).))))....)))).. ( -36.20) >DroVir_CAF1 2475 106 + 1 AUUCAUACA-CAUA-UUUGAAC-CGAUUAU-----------GUUUGCAGCUCGCGGUCAUGCUGCCAAGGCAGCCGCUAAGGCGGCCGCUGUAGCAAAUGGCAAAAUUGUAGCCGUAAUU .((((....-....-..)))).-.((((((-----------((((((.((..((((((..(((((....))))).((....)))))))).)).))))))(((.........))))))))) ( -35.50) >DroGri_CAF1 7076 105 + 1 AUUGACACA-GAUA-----AAC-UGAUUUU--------CUUUUUUGCAGCUCGCGGUCAUGCCGCCAAGGCAGCUGCUAAGGCGGCUGCUGGAGCCAAUGGCAAGAUUGUGGCUGUAAUU .......((-(...-----..)-)).....--------.....(((((((.(((((((.((((((...(((((((((....)))))))))...))....)))).)))))))))))))).. ( -44.80) >DroEre_CAF1 1124 116 + 1 ACUUAGAUG-CAUUUUGUAAAACUGAUCAAGCAACUGCUGCUUUUUCAGCUCGUAGCCAUGCUGCUAAGG---CUGCAAAGGCUGCGGCUGCCGCAAAUGGUAAGAUUGUGGCCGUAAUU .......((-(.....)))...((((..(((((.....)))))..))))..(((((((.(((.((....)---).)))..)))))))((.(((((((.........))))))).)).... ( -37.30) >DroMoj_CAF1 2108 105 + 1 --UCAAACUGAAUU-UUUUUAA-UGACUUA-----------UUUUCCAGCUCGCGGUCAUGCUGCCAAAGCUGCUGUUAAGGCGGCCGCUGGAGGAAAUGGAAAAAUUGUGGCCGUCAUU --............-.....((-((((...-----------(((((((((..((((.....))))....((((((.....)))))).)))))))))...((..(.....)..)))))))) ( -31.70) >DroPer_CAF1 522 99 + 1 AUUU-------AAUUUUUGAAAC--------------CAUCUUUUUUAGCUCGCAGCCAUGCCGCUAAGGCAGCUGUCAAGGCAGCCUCAGUUGCUAACGGAAAGAUUGUUGCGGUAAUU ....-------((((.(((.(((--------------.((((((((......(((((..((((.....))))(((((....)))))....)))))....)))))))).))).))).)))) ( -26.90) >consensus AUUUAAACA_CAUU_UUUAAAA_UGAUUAA__________CUUUUUCAGCUCGCAGCCAUGCUGCCAAGGCAGCUGCUAAGGCGGCCGCUGCAGCAAAUGGAAAGAUUGUGGCCGUAAUU ..............................................((((..(((((...))))).......(((((....))))).))))......((((...........)))).... (-13.47 = -13.28 + -0.19)
| Location | 1,053,147 – 1,053,264 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -25.57 |
| Energy contribution | -24.72 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
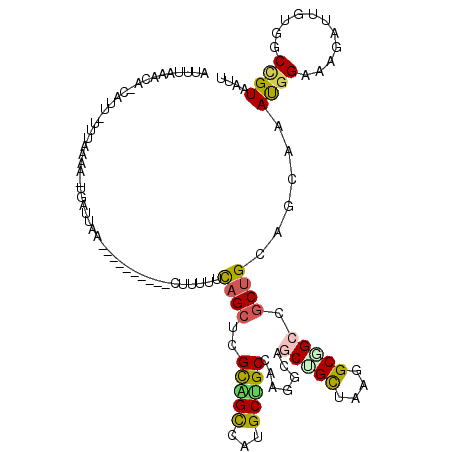
>4_DroMel_CAF1 1053147 117 + 1281640 CGUUUUCAGCUCGUAGCCAUGCUGCCAAGG---CUGCAAAGGCUGCAGCUGCCGCAAAUGGAAAGAUUGUGGCCGUAAUUGGUGCAGUCGUCGAUGUCCAGUUUGAUGAUAACUUACCGC .......((((.((((((.(((.((....)---).)))..))))))))))(((((((.(....)..))))))).......((((..((((((((........))))))))....)))).. ( -41.50) >DroVir_CAF1 2502 119 + 1 -GUUUGCAGCUCGCGGUCAUGCUGCCAAGGCAGCCGCUAAGGCGGCCGCUGUAGCAAAUGGCAAAAUUGUAGCCGUAAUUGGUGCCGUCGUGGACGUGCAAUUUGAUGAUAACUUGCCGC -((((((.((..((((((..(((((....))))).((....)))))))).)).))))))(((((.......((((....))))((((((...)))).))..............))))).. ( -46.10) >DroPse_CAF1 1 119 + 1 -UUUUUUAGCUCGCAGCCAUGCCGCUAAGGCAGCUGUCAAGGCAGCCUCAGUUGCUAACGGAAAGAUUGUUGCGGUAAUUGGUGCUGUCGUUGAUGUUCAGUUCGAUGAUGACUUGCCAC -....(((((..(((....))).)))))(((((..((((.((((((.((((((((((((((.....)))))..))))))))).))))))(((((........)))))..))))))))).. ( -40.40) >DroGri_CAF1 7101 120 + 1 UUUUUGCAGCUCGCGGUCAUGCCGCCAAGGCAGCUGCUAAGGCGGCUGCUGGAGCCAAUGGCAAGAUUGUGGCUGUAAUUGGUGCCGUCGUUGACGUGCAAUUCGAUGAUAACUUGCCAC ...(((((((.(((((((.((((((...(((((((((....)))))))))...))....)))).)))))))))))))).(((((..((((((((........))))))))....))))). ( -53.20) >DroWil_CAF1 1 113 + 1 -------AGCUCGAGGACAUGCCGCUAAGGCAGCUGCCAAGGCUGUCGCUGGAGGCAACGGCAAAAUUGUCGCUGUCAUUGGUGCUGUCGUCGAUGUACAGUUUGACGAUAAUUUGCCUC -------..(((.((((((.(((.....(((....)))..))))))).)).))).....((((((((((((((((((((((..(....)..))))).)))))..))))))..)))))).. ( -44.70) >DroMoj_CAF1 2134 119 + 1 -UUUUCCAGCUCGCGGUCAUGCUGCCAAAGCUGCUGUUAAGGCGGCCGCUGGAGGAAAUGGAAAAAUUGUGGCCGUCAUUGGUGCCGUCGUUGAUGUGCAGUUCGACGACAAUCUGCCAC -(((((((((..((((.....))))....((((((.....)))))).)))))))))............(((((.......(....)((((((((........)))))))).....))))) ( -44.10) >consensus _UUUUGCAGCUCGCGGUCAUGCCGCCAAGGCAGCUGCUAAGGCGGCCGCUGGAGCAAAUGGAAAAAUUGUGGCCGUAAUUGGUGCCGUCGUUGAUGUGCAGUUCGAUGAUAACUUGCCAC ............(((((..((((.....)))))))))...((((((.((((.......)))).........((((....)))))))((((((((........)))))))).....))).. (-25.57 = -24.72 + -0.86)

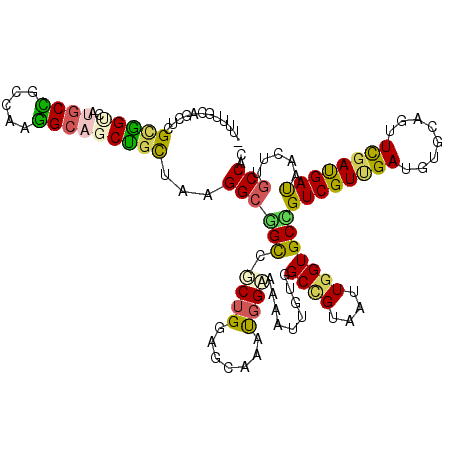
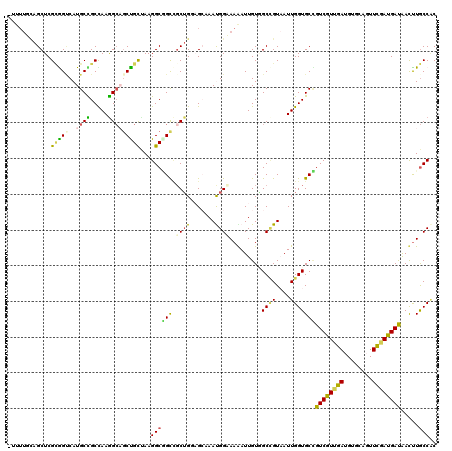
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:45 2006